Abstract
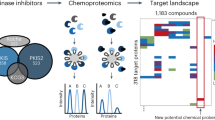
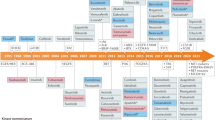
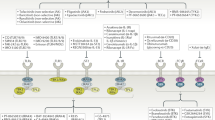
Despite the success of protein kinase inhibitors as approved therapeutics, drug discovery has focused on a small subset of kinase targets. Here we provide a thorough characterization of the Published Kinase Inhibitor Set (PKIS), a set of 367 small-molecule ATP-competitive kinase inhibitors that was recently made freely available with the aim of expanding research in this field and as an experiment in open-source target validation. We screen the set in activity assays with 224 recombinant kinases and 24 G protein–coupled receptors and in cellular assays of cancer cell proliferation and angiogenesis. We identify chemical starting points for designing new chemical probes of orphan kinases and illustrate the utility of these leads by developing a selective inhibitor for the previously untargeted kinases LOK and SLK. Our cellular screens reveal compounds that modulate cancer cell growth and angiogenesis in vitro. These reagents and associated data illustrate an efficient way forward to increasing understanding of the historically untargeted kinome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Bamborough, P. System-based drug discovery within the human kinome. Expert Opin. Drug Discov. 7, 1053–1070 (2012).
Cohen, P. & Alessi, D.R. Kinase drug discovery—what's next in the field? ACS Chem. Biol. 8, 96–104 (2013).
Wu, P., Nielsen, T.E. & Clausen, M.H. FDA-approved small-molecule kinase inhibitors. Trends Pharmacol. Sci. 36, 422–439 (2015).
Fedorov, O., Müller, S. & Knapp, S. The (un)targeted cancer kinome. Nat. Chem. Biol. 6, 166–169 (2010).
Edwards, A.M., Bountra, C., Kerr, D.J. & Willson, T.M. Open access chemical and clinical probes to support drug discovery. Nat. Chem. Biol. 5, 436–440 (2009).
Drewry, D.H., Willson, T.M. & Zuercher, W.J. Seeding collaborations to advance kinase science with the GSK Published Kinase Inhibitor Set (PKIS). Curr. Top. Med. Chem. 14, 340–342 (2014).
Zhang, J., Yang, P.L. & Gray, N.S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28–39 (2009).
Fedorov, O., Niesen, F.H. & Knapp, S. Kinase inhibitor selectivity profiling using differential scanning fluorimetry. Methods Mol. Biol. 795, 109–118 (2012).
Bamborough, P., Drewry, D., Harper, G., Smith, G.K. & Schneider, K. Assessment of chemical coverage of kinome space and its implications for kinase drug discovery. J. Med. Chem. 51, 7898–7914 (2008).
Witherington, J. et al. 5-aryl-pyrazolo[3,4-b]pyridazines: potent inhibitors of glycogen synthase kinase-3 (GSK-3). Bioorg. Med. Chem. Lett. 13, 1581–1584 (2003).
Conway, J.G. et al. Inhibition of colony-stimulating-factor-1 signaling in vivo with the orally bioavailable cFMS kinase inhibitor GW2580. Proc. Natl. Acad. Sci. USA 102, 16078–16083 (2005).
Takle, A.K. et al. The identification of potent and selective imidazole-based inhibitors of B-Raf kinase. Bioorg. Med. Chem. Lett. 16, 378–381 (2006).
Workman, P. & Collins, I. Probing the probes: fitness factors for small molecule tools. Chem. Biol. 17, 561–577 (2010).
Jester, B.W. et al. A coiled-coil enabled split-luciferase three-hybrid system: applied toward profiling inhibitors of protein kinases. J. Am. Chem. Soc. 132, 11727–11735 (2010).
Paolini, G.V., Shapland, R.H., van Hoorn, W.P., Mason, J.S. & Hopkins, A.L. Global mapping of pharmacological space. Nat. Biotechnol. 24, 805–815 (2006).
Ciceri, P. et al. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat. Chem. Biol. 10, 305–312 (2014).
Huber, K.V. et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature 508, 222–227 (2014).
Fourches, D. & Tropsha, A. Using graph indices for the analysis and comparison of chemical datasets. Mol. Inform. 32, 827–842 (2013).
Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 6, 813–823 (2006).
Weinstein, J.N. et al. An information-intensive approach to the molecular pharmacology of cancer. Science 275, 343–349 (1997).
Reinhold, W.C. et al. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 72, 3499–3511 (2012).
Miyazaki, Y. et al. Orally active 4-amino-5-diarylurea-furo[2,3-d]pyrimidine derivatives as anti-angiogenic agent inhibiting VEGFR2 and Tie-2. Bioorg. Med. Chem. Lett. 17, 1773–1778 (2007).
Ardini, E. et al. The TPM3-NTRK1 rearrangement is a recurring event in colorectal carcinoma and is associated with tumor sensitivity to TRKA kinase inhibition. Mol. Oncol. 8, 1495–1507 (2014).
Hasegawa, M. et al. Discovery of novel benzimidazoles as potent inhibitors of TIE-2 and VEGFR-2 tyrosine kinase receptors. J. Med. Chem. 50, 4453–4470 (2007).
Belkina, N.V., Liu, Y., Hao, J.J., Karasuyama, H. & Shaw, S. LOK is a major ERM kinase in resting lymphocytes and regulates cytoskeletal rearrangement through ERM phosphorylation. Proc. Natl. Acad. Sci. USA 106, 4707–4712 (2009).
Kuramochi, S. et al. LOK is a novel mouse STE20-like protein kinase that is expressed predominantly in lymphocytes. J. Biol. Chem. 272, 22679–22684 (1997).
Viswanatha, R., Ohouo, P.Y., Smolka, M.B. & Bretscher, A. Local phosphocycling mediated by LOK/SLK restricts ezrin function to the apical aspect of epithelial cells. J. Cell Biol. 199, 969–984 (2012).
Folkman, J. Angiogenesis: an organizing principle for drug discovery? Nat. Rev. Drug Discov. 6, 273–286 (2007).
Arnaoutova, I. & Kleinman, H.K. In vitro angiogenesis: endothelial cell tube formation on gelled basement membrane extract. Nat. Protoc. 5, 628–635 (2010).
Faivre, S., Demetri, G., Sargent, W. & Raymond, E. Molecular basis for sunitinib efficacy and future clinical development. Nat. Rev. Drug Discov. 6, 734–745 (2007).
Wilhelm, S. et al. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat. Rev. Drug Discov. 5, 835–844 (2006).
Olsson, A.K., Dimberg, A., Kreuger, J. & Claesson-Welsh, L. VEGF receptor signalling—in control of vascular function. Nat. Rev. Mol. Cell Biol. 7, 359–371 (2006).
Verheul, H.M. & Pinedo, H.M. Possible molecular mechanisms involved in the toxicity of angiogenesis inhibition. Nat. Rev. Cancer 7, 475–485 (2007).
Gao, Y. et al. A broad activity screen in support of a chemogenomic map for kinase signalling research and drug discovery. Biochem. J. 451, 313–328 (2013).
Anastassiadis, T., Deacon, S.W., Devarajan, K., Ma, H. & Peterson, J.R. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1039–1045 (2011).
Davis, M.I. et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1046–1051 (2011).
Fedorov, O. et al. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc. Natl. Acad. Sci. USA 104, 20523–20528 (2007).
Bain, J. et al. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 (2007).
Davies, S.P., Reddy, H., Caivano, M. & Cohen, P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351, 95–105 (2000).
Fabian, M.A. et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 23, 329–336 (2005).
Karaman, M.W. et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 26, 127–132 (2008).
Metz, J.T. et al. Navigating the kinome. Nat. Chem. Biol. 7, 200–202 (2011).
Posy, S.L. et al. Trends in kinase selectivity: insights for target class-focused library screening. J. Med. Chem. 54, 54–66 (2011).
Dranchak, P. et al. Profile of the GSK published protein kinase inhibitor set across ATP-dependent and-independent luciferases: implications for reporter-gene assays. PLoS One 8, e57888 (2013).
Atkinson, J.M. et al. An integrated in vitro and in vivo high-throughput screen identifies treatment leads for ependymoma. Cancer Cell 20, 384–399 (2011).
Ember, S.W. et al. Acetyl-lysine binding site of bromodomain-containing protein 4 (BRD4) interacts with diverse kinase inhibitors. ACS Chem. Biol. 9, 1160–1171 (2014).
Guo, K., Shelat, A.A., Guy, R.K. & Kastan, M.B. Development of a cell-based, high-throughput screening assay for ATM kinase inhibitors. J. Biomol. Screen. 19, 538–546 (2014).
Knight, Z.A. & Shokat, K.M. Features of selective kinase inhibitors. Chem. Biol. 12, 621–637 (2005).
Haystead, T.A. The purinome, a complex mix of drug and toxicity targets. Curr. Top. Med. Chem. 6, 1117–1127 (2006).
Besnard, J. et al. Automated design of ligands to polypharmacological profiles. Nature 492, 215–220 (2012).
Huang, X.P. et al. Parallel functional activity profiling reveals valvulopathogens are potent 5-hydroxytryptamine2B receptor agonists: implications for drug safety assessment. Mol. Pharmacol. 76, 710–722 (2009).
Wacker, D. et al. Structural features for functional selectivity at serotonin receptors. Science 340, 615–619 (2013).
Holbeck, S.L., Collins, J.M. & Doroshow, J.H. Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol. Cancer Ther. 9, 1451–1460 (2010).
Boutros, M., Brás, L.P. & Huber, W. Analysis of cell-based RNAi screens. Genome Biol. 7, R66 (2006).
Acknowledgements
The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Innovative Medicines Initiative (EU/EFPIA), Janssen, Merck & Co., Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome Trust. A.A.H.Z. acknowledges support from the BHF Centre of Research Excellence, Oxford (RE/13/1/30181).
Author information
Authors and Affiliations
Contributions
K.E.L., J.M.E., A.A.H.Z., J. Morris, S.R., N.S., B.A.T., B.L.R., D.H.D., S.K. and W.J.Z. designed the study. M.S., V.F., K.R.A.A., J.M.E., E.S., A.A.H.Z., J. Mikolajczyk, S.R., N.S. and X.-P.H. performed experiments. J.M.E., A.A.H.Z., J. Morris, B.A.T., A.T., X.-P.H., D.F., E.M., F.L.A., J.P.O., P.B., D.H.D., S.M., S.K., E.P., M.K. and W.J.Z. analyzed data. J.M.E., B.L.R., X.-P.H., A.A.H.Z., P.B., D.H.D., D.J.P., T.M.W., S.K. and W.J.Z. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.R. and N.S. are current employees of Nanosyn, Inc. J.P.O. is a current employee of Stratified Medical Ltd.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1-11 and Supplementary Figures 1-12 (PDF 6677 kb)
Rights and permissions
About this article
Cite this article
Elkins, J., Fedele, V., Szklarz, M. et al. Comprehensive characterization of the Published Kinase Inhibitor Set. Nat Biotechnol 34, 95–103 (2016). https://doi.org/10.1038/nbt.3374
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3374
This article is cited by
-
Identification of druggable regulators of cell secretion via a kinome-wide screen and high-throughput immunomagnetic cell sorting
Nature Biomedical Engineering (2023)
-
Chemogenetics for cell-type-specific modulation of signalling and neuronal activity
Nature Reviews Methods Primers (2023)
-
Opposing roles of CLK SR kinases in controlling HIV-1 gene expression and latency
Retrovirology (2022)
-
Dissecting the roles of Haspin and VRK1 in histone H3 phosphorylation during mitosis
Scientific Reports (2022)
-
A novel graph convolutional neural network for predicting interaction sites on protein kinase inhibitors in phosphorylation
Scientific Reports (2022)