Abstract
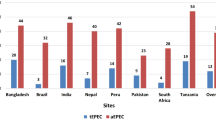
Enteropathogenic Escherichia coli (EPEC) are diarrhoeagenic E. coli, and are a significant cause of gastrointestinal illness among young children in developing countries. Typical EPEC are identified by the presence of the bundle-forming pilus encoded by a virulence plasmid, which has been linked to an increased severity of illness, while atypical EPEC lack this feature. Comparative genomics of 70 total EPEC from lethal (LI), non-lethal symptomatic (NSI) or asymptomatic (AI) cases of diarrhoeal illness in children enrolled in the Global Enteric Multicenter Study was used to investigate the genomic differences in EPEC isolates obtained from individuals with various clinical outcomes. A comparison of the genomes of isolates from different clinical outcomes identified genes that were significantly more prevalent in EPEC isolates of symptomatic and lethal outcomes than in EPEC isolates of asymptomatic outcomes. These EPEC isolates exhibited previously unappreciated phylogenomic diversity and combinations of virulence factors. These comparative results highlight the diversity of the pathogen, as well as the complexity of the EPEC virulence factor repertoire.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
118,99 € per year
only 9,92 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ochoa, T. J. & Contreras, C. A. Enteropathogenic Escherichia coli infection in children. Curr. Opin. Infect. Dis. 24, 478–483 (2011).
Kotloff, K. L. et al. The global enteric multicenter study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin. Infect. Dis. 55(Suppl 4), S232–S245 (2012).
Kotloff, K. L. et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209–222 (2013).
McDaniel, T. K., Jarvis, K. G., Donnenberg, M. S. & Kaper, J. B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl Acad. Sci. USA 92, 1664–1668 (1995).
Elliott, S. J. et al. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28, 1–4 (1998).
Perna, N. T. et al. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66, 3810–3817 (1998).
Tauschek, M., Strugnell, R. A. & Robins-Browne, R. M. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44, 1533–1550 (2002).
Kaper, J. B., Nataro, J. P. & Mobley, H. L. Pathogenic Escherichia coli. Nature Rev. Microbiol. 2, 123–140 (2004).
Tarr, P. I., Gordon, C. A. & Chandler, W. L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365, 1073–1086 (2005).
Clements, A., Young, J. C., Constantinou, N. & Frankel, G. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes 3, 71–87 (2012).
Pennington, H. Escherichia coli O157. Lancet 376, 1428–1435 (2010).
Nataro, J. P., Scaletsky, I. C., Kaper, J. B., Levine, M. M. & Trabulsi, L. R. Plasmid-mediated factors conferring diffuse and localized adherence of enteropathogenic Escherichia coli. Infect. Immun. 48, 378–383 (1985).
Bieber, D. et al. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280, 2114–2118 (1998).
Donnenberg, M. S., Giron, J. A., Nataro, J. P. & Kaper, J. B. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol. Microbiol. 6, 3427–3437 (1992).
Stone, K. D., Zhang, H. Z., Carlson, L. K. & Donnenberg, M. S. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol. Microbiol. 20, 325–337 (1996).
Donnenberg, M. S., Zhang, H. Z. & Stone, K. D. Biogenesis of the bundle-forming pilus of enteropathogenic Escherichia coli: reconstitution of fimbriae in recombinant E. coli and role of DsbA in pilin stability—a review. Gene 192, 33–38 (1997).
Nataro, J. P. & Kaper, J. B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201 (1998).
Hazen, T. H. et al. Refining the pathovar paradigm via phylogenomics of the attaching and effacing Escherichia coli. Proc. Natl Acad. Sci. USA 110, 12810–12815 (2013).
Tennant, S. M. et al. Characterisation of atypical enteropathogenic E. coli strains of clinical origin. BMC Microbiol. 9, 117 (2009).
Lacher, D. W., Steinsland, H., Blank, T. E., Donnenberg, M. S. & Whittam, T. S. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J. Bacteriol. 189, 342–350 (2007).
Rasko, D. A. et al. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190, 6881–6893 (2008).
Iguchi, A. et al. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191, 347–354 (2009).
Panchalingam, S. et al. Diagnostic microbiologic methods in the GEMS-1 case/control study. Clin. Infect. Dis. 55(Suppl 4), S294–S302 (2012).
Tenaillon, O., Skurnik, D., Picard, B. & Denamur, E. The population genetics of commensal Escherichia coli. Nature Rev. Microbiol. 8, 207–217 (2010).
Deng, W. et al. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl Acad. Sci. USA 101, 3597–3602 (2004).
Blank, T. E., Zhong, H., Bell, A. L., Whittam, T. S. & Donnenberg, M. S. Molecular variation among type IV pilin (bfpA) genes from diverse enteropathogenic Escherichia coli strains. Infect. Immun. 68, 7028–7038 (2000).
Rasko, D. A., Myers, G. S. & Ravel, J. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinf. 6, 2 (2005).
Sahl, J. W., Caporaso, J. G., Rasko, D. A. & Keim, P. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ 2, e332 (2014).
Sahl, J. W. et al. Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS ONE 8, e54287 (2013).
Touchon, M. et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5, e1000344 (2009).
Conner, C. P., Heithoff, D. M., Julio, S. M., Sinsheimer, R. L. & Mahan, M. J. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl Acad. Sci. USA 95, 4641–4645 (1998).
Heithoff, D. M. et al. Coordinate intracellular expression of Salmonella genes induced during infection. J. Bacteriol. 181, 799–807 (1999).
Hazen, T. H. et al. RNA-Seq analysis of isolate- and growth phase-specific differences in the global transcriptomes of enteropathogenic Escherichia coli prototype isolates. Front. Microbiol. 6, 569 (2015).
Donnenberg, M. S. et al. Bacterial factors associated with lethal outcome of enteropathogenic Escherichia coli infection: genomic case-control studies. PLoS Negl. Trop. Dis. 9, e0003791 (2015).
Sahl, J. W. et al. A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect. Immun. 79, 950–960 (2011).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Orvis, J. et al. Ergatis: a web interface and scalable software system for bioinformatics workflows. Bioinformatics 26, 1488–1492 (2010).
Galens, K. et al. The IGS standard operating procedure for automated prokaryotic annotation. Stand. Genomic Sci. 4, 244–251 (2011).
Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA 95, 14863–14868 (1998).
Saeed, A. I. et al. TM4 microarray software suite. Methods Enzymol. 411, 134–193 (2006).
R Development Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2010).
Angiuoli, S. V. & Salzberg, S. L. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27, 334–342 (2011).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Acknowledgements
This project was funded by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services NIH grant no. U19 AI090873.
Author information
Authors and Affiliations
Contributions
T.H.H., M.S.D., J.P.N. and D.A.R. conceived and designed the experiments. T.H.H., M.S.D., E.M.B., J.B.K., J.P.N. and D.A.R. performed the experiments. T.H.H., M.S.D., E.M.B., J.B.K., J.P.N. and D.A.R. analysed the data. S.P., M.A., A.H., I.M., J.B.O., T.R., B.T., S.Q., F.Q., A.Z., K.L.K., M.M.L. and J.P.N. contributed materials from the GEMS studies. T.H.H., M.S.D., E.M.B., J.B.K., J.P.N. and D.A.R. co-wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Results, Figures 1–6, References and Tables 1–10 (PDF 8881 kb)
Supplementary Data 1
Supplementary Data 1 (TXT 10399 kb)
Supplementary Data 2
Supplementary Data 2 (TXT 9913 kb)
Rights and permissions
About this article
Cite this article
Hazen, T., Donnenberg, M., Panchalingam, S. et al. Genomic diversity of EPEC associated with clinical presentations of differing severity. Nat Microbiol 1, 15014 (2016). https://doi.org/10.1038/nmicrobiol.2015.14
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2015.14
This article is cited by
-
Genomic diversity of non-diarrheagenic fecal Escherichia coli from children in sub-Saharan Africa and south Asia and their relatedness to diarrheagenic E. coli
Nature Communications (2023)
-
Human gut-derived B. longum subsp. longum strains protect against aging in a d-galactose-induced aging mouse model
Microbiome (2021)
-
The population genetics of pathogenic Escherichia coli
Nature Reviews Microbiology (2021)
-
Diversity among blaKPC-containing plasmids in Escherichia coli and other bacterial species isolated from the same patients
Scientific Reports (2018)
-
Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life
Nature Microbiology (2018)