Abstract
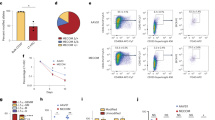
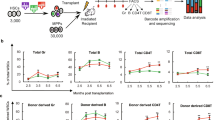
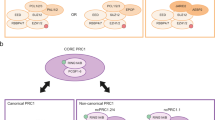
Polycomb group (PcG) proteins play important roles in hematopoietic stem cell (HSC) self-renewal. Mel18 and Bmi1 are homologs of the PCGF subunit within the Polycomb repressive complex 1 (PRC1). Bmi1 (PCGF4) enhances HSC self-renewal and promotes terminal differentiation. However, the role of Mel18 (PCGF2) in hematopoiesis is not fully understood and how Mel18 regulates gene transcription in HSCs remains elusive. We found that acute deletion of Mel18 in the hematopoietic compartment significantly increased the frequency of functional HSCs in the bone marrow. Furthermore, we demonstrate that Mel18 inhibits HSC self-renewal and proliferation. RNA-seq studies revealed that HSC self-renewal and proliferation gene signatures are enriched in Mel18−/− hematopoietic stem and progenitors (HSPCs) compared to Mel18+/+ HSPCs. Notably, ATAC-seq revealed increased chromatin accessibility at genes important for HSC self-renewal, whereas CUT&RUN showed decreased enrichment of H2AK119ub1 at genes important for proliferation, leading to increased expression of both Hoxb4 and Cdk4 in Mel18−/− HSPCs. Thus, we demonstrate that Mel18 inhibits hematopoietic stem cell self-renewal through repressing the transcription of genes important for HSC self-renewal and proliferation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Normalized sequencing data can be accessed from the Gene Expression Omnibus (GSE279249, GSE279250, GSE279251 and GSE279252). Requests for raw sequencing data should be addressed to and will be fulfilled by the corresponding author (YL).
Code availability
No custom code or software was used as part of the data analysis. All packages used are listed in the “Methods” section.
References
Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806.
Attar EC, Scadden DT. Regulation of hematopoietic stem cell growth. Leukemia 2004;18:1760–8.
Sorrentino BP. Clinical strategies for expansion of haematopoietic stem cells. Nat Rev Immunol. 2004;4:878–88.
Walasek MA, van Os R, de Haan G. Hematopoietic stem cell expansion: challenges and opportunities. Ann N Y Acad Sci. 2012;1266:138–50.
Akala OO, Clarke MF. Hematopoietic stem cell self-renewal. Curr Opin Genet Dev. 2006;16:496–501.
Zon LI. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature 2008;453:306–13.
Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–84.
Piunti A, Shilatifard A. The roles of Polycomb repressive complexes in mammalian development and cancer. Nat Rev Mol Cell Biol. 2021;22:326–45.
Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, et al. Pcgf homologs, Cbx proteins, and Rybp define functionally distinct Prc1 family complexes. Mol Cell. 2012;45:344–56.
Lessard J, Sauvageau G. Polycomb group genes as epigenetic regulators of normal and leukemic hemopoiesis. Exp Hematol. 2003;31:567–85.
Konuma T, Oguro H, Iwama A. Role of the polycomb group proteins in hematopoietic stem cells. Dev Growth Differ. 2010;52:505–16.
Vidal M, Starowicz K. Polycomb complexes PRC1 and their function in hematopoiesis. Exp Hematol. 2017;48:12–31.
Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–5.
Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, Tsukui H, et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21:843–51.
Liu Y, Liu F, Yu H, Zhao X, Sahsida G, Deblasio A, et al. Akt phosphorylates the transcriptional repressor Bmi1 to block its association with tumor suppressing Ink4a-Arf locus. Sci Signal. 2012;5:ra77.
Yu H, Gao R, Chen S, Liu X, Wang Q, Cai W, et al. Bmi1 regulates Wnt signaling in hematopoietic stem and progenitor cells. Stem Cell Rev Rep. 2021;17:2304–13.
Gao R, Chen S, Kobayashi M, Yu H, Young SK, Soltis A, et al. Bmi1 promotes erythroid development through regulating ribosome biogenesis. Stem Cells. 2015;33:925–38.
Kobayashi M, Lin Y, Mishra A, Shelly C, Gao R, Wang P, et al. Bmi1 maintains the self-renewal property of innate -like B lymphocyes. J Immunol. 2020;204:3262–72.
Kajiume T, Ninomiya Y, Ishihara H, Kanno R, Kanno M. Polycomb group gene Mel-18 modulates the self-renewal activity and cell cycle status of hematopoietic stem cells. Exp Hematol. 2004;32:571–8.
Oguro H, Yuan J, Ichikawa H, Ikawa T, Yamazaki S, Kawamoto H, et al. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell. 2010;6:279–86.
Liu X, Wei W, Li X, Shen P, Ju D, Wang Z, et al. BMI1 and MEL18 promote colitis-associated cancer in mice via REG3B and STAT3. Gastroenterology. 2017;153:1607–20.
Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, Huang G, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4:37–48.
Chen S, Gao R, Yao C, Kobayashi M, Liu SZ, Yoder MC, et al. Genotoxic stresses promotes the clonal expansion of hematopoietic stem cells expressing mutant p53. Leukemia. 2018;32:850–4.
Chen S, Wang Q, Yu H, Capitano ML, Vemula S, Nabinger SC, et al. Mutant p53 drives clonal hematopoiesis through modulating epigenetic pathway. Nat Commun. 2019;10:5649.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–9.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
Liu T. Use model-based analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol Biol. 2014;1150:81–95.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinforma. 2011;12:35.
Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, et al. ChIPpeakAnno: a bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinforma. 2010;11:237.
Levine SL, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–8.
Parreno V, Martinez AM, Cavalli G. Mechanisms of polycomb group protein function in cancer. Cell Res. 2022;32:231–53.
Grubach L, Juhl-Christensen C, Rethmeier A, Olesen LH, Aggerholm A, Hokland P, et al. Gene expression profiling of Polycomb, Hox and Meis genes in patients with acute myeloid leukaemia. Eur J Haematol. 2008;81:112–22.
Antonchuk J, Sauvageau G, Humphries RK. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell. 2002;109:39–45.
Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and polycomb-group gene Bmi-1 regulates cell proliferation and senescence through the Ink4a Locus. Nature. 1999;397:164–8.
Oguro H, Iwama A, Morita Y, Kamijo T, van Lohuizen M, Nakauchi H. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J Exp Med. 2006;203:2247–53.
Yu M, Mazor T, Huang H, Huang HT, Kathrein K, Woo AJ, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell. 2012;45:330–43.
Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8.
Tamburri S, Lavarone E, Fernández-Pérez D, Conway E, Zanotti M, Manganaro D, et al. Histone H2AK119 mono-ubiquitination is essential for polycomb-mediated transcriptional repression. Mol Cell. 2020;77:840–56.e5.
Kajiume T, Ohno N, Sera Y, Kawahara Y, Yuge L, Kobayashi M. Reciprocal expression of Bmi1 and Mel-18 is associated with functioning of primitive hematopoietic cells. Exp Hematol. 2009;37:857–66.e2.
Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37.
Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–60.
Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–54.
Scelfo A, Fernández-Pérez D, Tamburri S, Zanotti M, Lavarone E, Soldi M, et al. Functional landscape of PCGF proteins reveals both RING1A/B-dependent-and RING1A/B-independent-specific activities. Mol Cell. 2019;74:1037–52.e7.
Fursova NA, Blackledge NP, Nakayama M, Ito S, Koseki Y, Farcas AM, et al. Synergy between variant PRC1 complexes defines polycomb-mediated gene repression. Mol Cell. 2019;74:1020–36.e8.
Acknowledgements
We would like to acknowledge the Flow Cytometry Core, Pathology Core, NU Seq core, Quantitative Data Science Core, and Center for Comparative Medicine at Northwestern University Feinberg School of Medicine and Robert H. Lurie Comprehensive Cancer Center. This work was supported by R01 HL150624, R56 DK119524, R56 AG052501, DoD W81XWH-18-1-0265, and DoD W81XWH-19-1-0575 awards (to YL). This work was supported in part by the Leukemia & Lymphoma Society (LLS) Translational Research Program award 6581-20 and the St. Baldrick’s Foundation Scholar Award (to YL). SC was supported by the LLS Career Development Special Fellow Award, American Society of Hematology (ASH) Research Restart Award, and Lady Tata Memorial International Awards for Research in Leukaemia. SB was supported by a NIH F31 Award F31HL160120.
Author information
Authors and Affiliations
Contributions
WC, SC, and YL designed the research. WC, XL, SB, SX, SV, HC, YY, CB, DH, YJ, MH, TMM, and SC performed the research; WC, SL, JW, FY, SC, and YL analyzed the data and performed the statistical analysis. MLC, HL, PJ, ZG, DP, LCP, and RX provided reagents and constructive advice to the study. WC, SC, and YL wrote the manuscript. All authors read, comment on, and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations. We have complied with all the relevant ethical regulations for animal testing and research. All animal-related experiments have received ethical approval from both Indiana and Northwestern University Institutional Animal Care and Use Committee (IACUC).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cai, W., Liu, X., Barajas, S. et al. Polycomb group protein Mel18 inhibits hematopoietic stem cell self-renewal through repressing the transcription of self-renewal and proliferation genes. Leukemia 39, 296–307 (2025). https://doi.org/10.1038/s41375-024-02462-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-024-02462-w