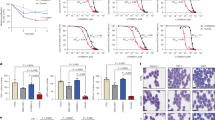
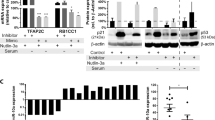
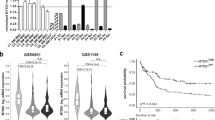
Abstract
Recent extensive studies on the genomic and molecular profiles of acute myeloid leukemia (AML) have expanded the treatment options, including, a range of compounds represented by fms-like tyrosine kinase 3 and isocitrate dehydrogenase 1/2 inhibitors. However, despite this progress, further treatments for AML are still required. Adenosine deaminase acting on RNA 1 (ADAR1) has been shown to play an important oncogenic role in many cancers, but its involvement in AML progression remains underexplored. In this study, we demonstrated that ADAR1 was overexpressed in AML and served as a crucial oncogenic target. Loss of ADAR1 inhibited the Wnt signaling pathway, blocked AML cell proliferation, and induced apoptosis. Importantly, we demonstrate that ADAR1, as an RNA-binding protein, interacts with pri-miR-766 independently of its editing function, regulating the maturation of miR-766-3p and enhancing the expression of WNT5B. Genetic inhibition or use of the ADAR1 inhibitor ZYS-1 significantly suppressed AML cell growth both in vitro and in vivo. Overall, these results elucidated the tumorigenic mechanism of ADAR1 and validated it as a potential drug target in AML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The raw sequence data (GSA-Human: HRA008848) reported in this paper have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human. All data generated during this study are included in this published article and its supplementary files. The AML data from the TCGA platform analyzed in this study are available at https://portal.gdc.cancer.gov/. The 70 normal tissue data from GTEx analyzed in his study are available at https://www.gtexportal.org. Source data from GSE34577 and GSE24395 analyzed in this study are available at https://www.ncbi.nlm.nih.gov/geo/. Data for miRNA analysis were obtained from sites TargetScan (https://www.targetscan.org/vert_80/) and miRDB (https://mirdb.org/).
Code availability
No custom code was created in this study.
References
DiNardo CD, Erba HP, Freeman SD, Wei AH. Acute myeloid leukaemia. Lancet. 2023;401:2073–86.
San Jose-Eneriz E, Gimenez-Camino N, Rabal O, Garate L, Miranda E, Gomez-Echarte N, et al. Epigenetic-based differentiation therapy for Acute Myeloid Leukemia. Nat Commun. 2024;15:5570.
Thol F, Dohner H, Ganser A. How I treat refractory and relapsed acute myeloid leukemia. Blood. 2024;143:11–20.
Jayavelu AK, Wolf S, Buettner F, Alexe G, Haupl B, Comoglio F, et al. The proteogenomic subtypes of acute myeloid leukemia. Cancer Cell. 2022;40:301–17.e12.
Stratmann S, Vesterlund M, Umer HM, Eshtad S, Skaftason A, Herlin MK, et al. Proteogenomic analysis of acute myeloid leukemia associates relapsed disease with reprogrammed energy metabolism both in adults and children. Leukemia. 2023;37:550–9.
Li JF, Cheng WY, Lin XJ, Wen LJ, Wang K, Zhu YM, et al. Aging and comprehensive molecular profiling in acute myeloid leukemia. Proc Natl Acad Sci USA. 2024;121:e2319366121.
Liu H. Emerging agents and regimens for AML. J Hematol Oncol. 2021;14:49.
Kayser S, Levis MJ. The clinical impact of the molecular landscape of acute myeloid leukemia. Haematologica. 2023;108:308–20.
Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019;33:299–312.
Barajas JM, Rasouli M, Umeda M, Hiltenbrand R, Abdelhamed S, Mohnani R, et al. Acute myeloid leukemias with UBTF tandem duplications are sensitive to menin inhibitors. Blood. 2024;143:619–30.
Decroocq J, Birsen R, Montersino C, Chaskar P, Mano J, Poulain L, et al. RAS activation induces synthetic lethality of MEK inhibition with mitochondrial oxidative metabolism in acute myeloid leukemia. Leukemia. 2022;36:1237–52.
Droegemeier K, Baselga J, Fouse J. BCL2/MDM2 inhibitor combo effective in AML. Cancer Discov. 2019;9:156.
Pan R, Ruvolo V, Mu H, Leverson JD, Nichols G, Reed JC, et al. Synthetic lethality of combined Bcl-2 inhibition and p53 activation in AML: mechanisms and superior antileukemic efficacy. Cancer Cell. 2017;32:748–60.e6.
Mill CP, Fiskus W, DiNardo CD, Birdwell C, Davis JA, Kadia TM, et al. Effective therapy for AML with RUNX1 mutation by cotreatment with inhibitors of protein translation and BCL2. Blood. 2022;139:907–21.
Newell LF, Cook RJ. Advances in acute myeloid leukemia. BMJ. 2021;375:n2026.
Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–3.
Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 2016;17:83–96.
Song B, Shiromoto Y, Minakuchi M, Nishikura K. The role of RNA editing enzyme ADAR1 in human disease. Wiley Interdiscip Rev RNA. 2022;13:e1665.
Baker AR, Slack FJ. ADAR1 and its implications in cancer development and treatment. Trends Genet. 2022;38:821–30.
Shigeyasu K, Okugawa Y, Toden S, Miyoshi J, Toiyama Y, Nagasaka T, et al. AZIN1 RNA editing confers cancer stemness and enhances oncogenic potential in colorectal cancer. JCI Insight. 2018;3(12):e99976.
Luo J, Gong L, Yang Y, Zhang Y, Liu Q, Bai L, et al. Enhanced mitophagy driven by ADAR1-GLI1 editing supports the self-renewal of cancer stem cells in hepatocellular carcinoma. Hepatology. 2024;79:61–78.
Teoh PJ, An O, Chung TH, Chooi JY, Toh SHM, Fan S, et al. Aberrant hyperediting of the myeloma transcriptome by ADAR1 confers oncogenicity and is a marker of poor prognosis. Blood. 2018;132:1304–17.
Jiang Q, Isquith J, Zipeto MA, Diep RH, Pham J, Delos Santos N, et al. Hyper-editing of cell-cycle regulatory and tumor suppressor RNA promotes malignant progenitor propagation. Cancer Cell. 2019;35:81–94.e7.
Jiao Y, Xu Y, Liu C, Miao R, Liu C, Wang Y, et al. The role of ADAR1 through and beyond its editing activity in cancer. Cell Commun Signal. 2024;22(1):42.
Meduri E, Breeze C, Marando L, Richardson SE, Huntly BJP. The RNA editing landscape in acute myeloid leukemia reveals associations with disease mutations and clinical outcome. iScience. 2022;25:105622.
Xiao H, Cheng Q, Wu X, Tang Y, Liu J, Li X. ADAR1 may be involved in the proliferation of acute myeloid leukemia cells via regulation of the Wnt pathway. Cancer Manag Res. 2019;11:8547–55.
Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–22.
Menon A, Abd-Aziz N, Khalid K, Poh CL, Naidu R. miRNA: a promising therapeutic target in cancer. Int J Mol Sci. 2022;23(19):11502.
Cho C, Myung S-J, Chang S. ADAR1 and MicroRNA, a hidden crosstalk in cancer. Int J Mol Sci. 2017;18(4):799.
Zipeto MA, Court AC, Sadarangani A, Delos Santos NP, Balaian L, Chun HJ, et al. ADAR1 activation drives leukemia stem cell self-renewal by impairing let-7 biogenesis. Cell Stem Cell. 2016;19:177–91.
Wang Y, Xu X, Yu S, Jeong KJ, Zhou Z, Han L, et al. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res. 2017;27:1112–25.
Zhang S, Chen H, Liu W, Fang L, Qian Z, Kong R, et al. miR-766-3p targeting BCL9L suppressed tumorigenesis, epithelial-mesenchymal transition, and metastasis through the beta-catenin signaling pathway in osteosarcoma cells. Front Cell Dev Biol. 2020;8:594135.
You Y, Que K, Zhou Y, Zhang Z, Zhao X, Gong J, et al. MicroRNA-766-3p inhibits tumour progression by targeting Wnt3a in hepatocellular carcinoma. Mol Cells. 2018;41:830–41.
Chen T, Xiang JF, Zhu SS, Chen SY, Yin QF, Zhang XO, et al. ADAR1 is required for differentiation and neural induction by regulating microRNA processing in a catalytically independent manner. Cell Res. 2015;25:459–76.
Quick-Cleveland J, Jacob JP, Weitz SH, Shoffner G, Senturia R, Guo F. The DGCR8 RNA-binding heme ___domain recognizes primary microRNAs by clamping the hairpin. Cell Rep. 2014;7:1994–2005.
Wang X, Li J, Zhu Y, Zeng T, Ding J, Min W, et al. Targeting ADAR1 with a novel small-molecule for the treatment of prostate cancer. Res. Sq. 2021;1–39. https://doi.org/10.21203/rs.3.rs-879741/v1.
Guo M, Chan THM, Zhou Q, An O, Li Y, Song Y, et al. Core-binding factor fusion downregulation of ADAR2 RNA editing contributes to AML leukemogenesis. Blood. 2023;141:3078–90.
Bahn JH, Ahn J, Lin X, Zhang Q, Lee JH, Civelek M, et al. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat Commun. 2015;6:6355.
Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21.
Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–9.
Acknowledgements
This study was supported by the National Key R&D Program of China (2022YFA1303803), National Natural Science Foundation of China (82373738, 82321005, 82304296), and the Project Program of State Key Laboratory of Natural Medicines, China Pharmaceutical University (SKLNMZZ2024JS05). The present study was also supported by Natural Science Foundation of Jiangsu Province (BK20240013, BK20231013), Jiangsu Funding Program for Excellent Postdoctoral Talent (2023ZB125), The Fundamental Research Funds for the Central Universities (2632024ZD06, 2632023GR22). Some images in this article were created using BioRender.com.
Author information
Authors and Affiliations
Contributions
PY and XW conceived the study. XW, Z-RS, and J-XL designed the research. Z-RS and J-XL performed most of the experiments with the help of J-YD, Y-WZ, W-JM, Y-SZ, YH, KY, C-LS, X-JW, HS, and L-PW. J-YD and Y-WZ conducted the bioinformatics analysis. All the authors analyzed the data. Z-RS, J-XL, J-YD, and XW wrote the manuscript, while PY, XW, W-BK, and S-QL reviewed and revised it. PY, XW, and W-BK supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, Z., Li, J., Ding, J. et al. ADAR1 is required for acute myeloid leukemia cell survival by modulating post-transcriptional Wnt signaling through impairing miRNA biogenesis. Leukemia 39, 599–613 (2025). https://doi.org/10.1038/s41375-024-02500-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-024-02500-7