Abstract
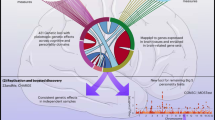
Individual variations of white matter (WM) tracts are known to be associated with various cognitive and neuropsychiatric traits. Diffusion tensor imaging (DTI) and genome-wide single-nucleotide polymorphism (SNP) data from 17,706 UK Biobank participants offer the opportunity to identify novel genetic variants of WM tracts and explore the genetic overlap with other brain-related complex traits. We analyzed the genetic architecture of 110 tract-based DTI parameters, carried out genome-wide association studies (GWAS), and performed post-GWAS analyses, including association lookups, gene-based association analysis, functional gene mapping, and genetic correlation estimation. We found that DTI parameters are substantially heritable for all WM tracts (mean heritability 48.7%). We observed a highly polygenic architecture of genetic influence across the genome (p value = 1.67 × 10−05) as well as the enrichment of genetic effects for active SNPs annotated by central nervous system cells (p value = 8.95 × 10−12). GWAS identified 213 independent significant SNPs associated with 90 DTI parameters (696 SNP-level and 205 locus-level associations; p value < 4.5 × 10−10, adjusted for testing multiple phenotypes). Gene-based association study prioritized 112 significant genes, most of which are novel. More importantly, association lookups found that many of the novel SNPs and genes of DTI parameters have previously been implicated with cognitive and mental health traits. In conclusion, the present study identifies many new genetic variants at SNP, locus and gene levels for integrity of brain WM tracts and provides the overview of pleiotropy with cognitive and mental health traits.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
We made use of publicly available software and tools. All codes used to generate results that are reported in this paper are available upon request.
References
Penke L, Maniega SM, Bastin M, Hernández MV, Murray C, Royle N, et al. Brain-wide white matter tract integrity is associated with information processing speed and general intelligence. Mol Psychiatry. 2012;17:955.
Tamnes CK, Østby Y, Walhovd KB, Westlye LT, Due‐Tønnessen P, Fjell AM. Intellectual abilities and white matter microstructure in development: a diffusion tensor imaging study. Hum Brain Mapp. 2010;31:1609–25.
Ritchie SJ, Bastin ME, Tucker-Drob EM, Maniega SM, Engelhardt LE, Cox SR, et al. Coupled changes in brain white matter microstructure and fluid intelligence in later life. J Neurosci. 2015;35:8672–82.
Ritchie SJ, Booth T, Hernández MdCV, Corley J, Maniega SM, Gow AJ, et al. Beyond a bigger brain: multivariable structural brain imaging and intelligence. Intelligence. 2015;51:47–56.
Nir TM, Jahanshad N, Villalon-Reina JE, Toga AW, Jack CR, Weiner MW, et al. Effectiveness of regional DTI measures in distinguishing Alzheimer’s disease, MCI, and normal aging. NeuroImage. 2013;3:180–95.
Bohnen NI, Albin RL. White matter lesions in Parkinson disease. Nat. Rev. Neurol. 2011;7:229–36.
Voineskos AN. Genetic underpinnings of white matter ‘connectivity’: heritability, risk, and heterogeneity in schizophrenia. Schizophr Res. 2015;161:50–60.
Sudre G, Choudhuri S, Szekely E, Bonner T, Goduni E, Sharp W, et al. Estimating the heritability of structural and functional brain connectivity in families affected by attention-deficit/hyperactivity disorder. JAMA Psychiatry. 2017;74:76–84.
Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson. 1994;103:247–54.
Beaulieu C. The basis of anisotropic water diffusion in the nervous system–a technical review. NMR Biomed. 2002;15:435–55.
Jones DK, Knösche TR, Turner R. White matter integrity, fiber count, and other fallacies: the do’s and don’ts of diffusion MRI. Neuroimage. 2013;73:239–54.
Cox SR, Ritchie SJ, Tucker-Drob EM, Liewald DC, Hagenaars SP, Davies G, et al. Ageing and brain white matter structure in 3,513 UK Biobank participants. Nat Commun. 2016;7:13629.
Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487–505.
Tamnes CK, Roalf DR, Goddings A-L, Lebel C. Diffusion MRI of white matter microstructure development in childhood and adolescence: methods, challenges and progress. Dev Cogn Neurosci. 2018;33:161–75.
Lee SJ, Steiner RJ, Luo S, Neale MC, Styner M, Zhu H, et al. Quantitative tract-based white matter heritability in twin neonates. Neuroimage. 2015;111:123–35.
Lee SJ, Steiner RJ, Yu Y, Short SJ, Neale MC, Styner MA, et al. Common and heritable components of white matter microstructure predict cognitive function at 1 and 2 y. Proc Natl Acad Sci USA. 2017;114:148–53.
Brouwer RM, Mandl RC, Peper JS, van Baal GCM, Kahn RS, Boomsma DI, et al. Heritability of DTI and MTR in nine-year-old children. Neuroimage. 2010;53:1085–92.
Chiang M-C, Barysheva M, Toga AW, Medland SE, Hansell NK, James MR, et al. BDNF gene effects on brain circuitry replicated in 455 twins. Neuroimage. 2011;55:448–54.
Kochunov P, Jahanshad N, Marcus D, Winkler A, Sprooten E, Nichols TE, et al. Heritability of fractional anisotropy in human white matter: a comparison of Human Connectome Project and ENIGMA-DTI data. Neuroimage. 2015;111:300–11.
Vuoksimaa E, Panizzon MS, Hagler DJ Jr, Hatton SN, Fennema‐Notestine C, Rinker D, et al. Heritability of white matter microstructure in late middle age: a twin study of tract‐based fractional anisotropy and absolute diffusivity indices. Hum Brain Mapp. 2017;38:2026–36.
Kanchibhotla SC, Mather KA, Wen W, Schofield PR, Kwok JB, Sachdev PS. Genetics of ageing-related changes in brain white matter integrity–A review. Ageing Res Rev. 2013;12:391–401.
Timpson NJ, Greenwood CM, Soranzo N, Lawson DJ, Richards JB. Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat Rev Genet. 2017;19:110–24.
Badano JL, Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nat Rev Genet. 2002;3:779–89.
Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–86.
Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82.
Loh P-R, Bhatia G, Gusev A, Finucane HK, Bulik-Sullivan BK, Pollack SJ, et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat Genet. 2015;47:1385–92.
Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–25.
Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–35.
Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, et al. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101:5–22.
Watanabe K, Taskesen E, Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826.
Watanabe K, Stringer S, Frei O, Mirkov MU, Polderman TJ, van der Sluis S, et al. A global view of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019;51:1339–48.
Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41.
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779.
Alfaro-Almagro F, Jenkinson M, Bangerter NK, Andersson JL, Griffanti L, Douaud G, et al. Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage. 2018;166:400–24.
Rutten-Jacobs LC, Tozer DJ, Duering M, Malik R, Dichgans M, Markus HS, et al. Genetic study of white matter integrity in UK Biobank (N = 8448) and the overlap with stroke, depression, and dementia. Stroke. 2018;49:1340–7.
Elliott LT, Sharp K, Alfaro-Almagro F, Shi S, Miller KL, Douaud G, et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 2018;562:210–6.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219.
Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2018;47:D1005–D1012.
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. Genome-wide genetic data on ~500,000 UK Biobank participants. Nature. 2018;562:203–9.
Jahanshad N, Kochunov PV, Sprooten E, Mandl RC, Nichols TE, Almasy L, et al. Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: a pilot project of the ENIGMA–DTI working group. Neuroimage. 2013;81:455–69.
Kochunov P, Jahanshad N, Sprooten E, Nichols TE, Mandl RC, Almasy L, et al. Multi-site study of additive genetic effects on fractional anisotropy of cerebral white matter: comparing meta and megaanalytical approaches for data pooling. Neuroimage. 2014;95:136–50.
Kim MJ, Elliott ML, d’Arbeloff TC, Knodt AR, Radtke SR, Brigidi BD, et al. Microstructural integrity of white matter moderates an association between childhood adversity and adult trait anger. Aggress Behav. 2019;45:310–8.
Kelly S, Jahanshad N, Zalesky A, Kochunov P, Agartz I, Alloza C, et al. Widespread white matter microstructural differences in schizophrenia across 4322 individuals: results from the ENIGMA schizophrenia DTI working group. Mol Psychiatry. 2018;23:1261–9.
Dennison MJ, Rosen ML, Sambrook KA, Jenness JL, Sheridan MA, McLaughlin KA. Differential associations of distinct forms of childhood adversity with neurobehavioral measures of reward processing: a developmental pathway to depression. Child Dev. 2019;90:e96–e113.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Consortium G. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60.
Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–28.
Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–53.
Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016;17:2042–59.
Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30.
Consortium IS. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52.
Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW. Polygenic prediction via bayesian regression and continuous shrinkage priors. Nat. Commun. 2019;10:1776.
Pasaniuc B, Price AL. Dissecting the genetics of complex traits using summary association statistics. Nat Rev Genet. 2017;18:117–27.
Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–21.
Nagel M, Jansen PR, Stringer S, Watanabe K, de Leeuw CA, Bryois J, et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet. 2018;50:920–27.
Davies G, Lam M, Harris SE, Trampush JW, Luciano M, Hill WD, et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat Commun. 2018;9:2098.
Luciano M, Hagenaars SP, Davies G, Hill WD, Clarke T-K, Shirali M, et al. Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nat Genet. 2018;50:6–11.
Kouri N, Ross OA, Dombroski B, Younkin CS, Serie DJ, Soto-Ortolaza A, et al. Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat Commun. 2015;6:7247.
Lam M, Trampush JW, Yu J, Knowles E, Davies G, Liewald DC, et al. Large-scale cognitive GWAS meta-analysis reveals tissue-specific neural expression and potential nootropic drug targets. Cell Rep. 2017;21:2597–613.
Chang D, Nalls MA, Hallgrímsdóttir IB, Hunkapiller J, van der Brug M, Cai F, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49:1511–6.
Okbay A, Baselmans BM, De Neve J-E, Turley P, Nivard MG, Fontana MA, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48:624–33.
Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, Team AR, Consortium SUDWGotPG, et al. Genome-wide association study meta-analysis of the Alcohol Use Disorders Identification Test (AUDIT) in two population-based cohorts. Am J Psychiatry. 2018;176:107–18.
Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert J-C, Chung J, Naj AC, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry. 2016;21:108–17.
Trampush JW, Yang M, Yu J, Knowles E, Davies G, Liewald D, et al. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol Psychiatry. 2017;22:336–45.
Li Z, Chen J, Yu H, He L, Xu Y, Zhang D, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49:1576–83.
Bergen S, O’dushlaine C, Ripke S, Lee P, Ruderfer D, Akterin S, et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry. 2012;17:880–6.
Kramer PL, Xu H, Woltjer RL, Westaway SK, Clark D, Erten-Lyons D, et al. Alzheimer disease pathology in cognitively healthy elderly: a genome-wide study. Neurobiol Aging. 2011;32:2113–22.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B.1995;57:289–300.
Satterthwaite TD, Elliott MA, Ruparel K, Loughead J, Prabhakaran K, Calkins ME, et al. Neuroimaging of the Philadelphia neurodevelopmental cohort. Neuroimage. 2014;86:544–53.
Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–13.
Clarke T, Lupton M, Fernandez-Pujals A, Starr J, Davies G, Cox S, et al. Common polygenic risk for autism spectrum disorder (ASD) is associated with cognitive ability in the general population. Mol Psychiatry. 2016;21:419–25.
Mistry S, Harrison JR, Smith DJ, Escott-Price V, Zammit S. The use of polygenic risk scores to identify phenotypes associated with genetic risk of bipolar disorder and depression: a systematic review. J Affect Disord. 2018;234:148–55.
Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14:483–95.
Bach M, Laun FB, Leemans A, Tax CM, Biessels GJ, Stieltjes B, et al. Methodological considerations on tract-based spatial statistics (TBSS). Neuroimage. 2014;100:358–69.
Acknowledgements
This research was partially supported by U.S. NIH grants MH086633 and MH116527, and a grant from the Cancer Prevention Research Institute of Texas. We thank the individuals represented in the UK Biobank and the Philadelphia Neurodevelopmental Cohort (PNC) datasets for their participation and the research teams for their work in collecting, processing and disseminating these datasets for analysis. This research has been conducted using the UK Biobank resource (application number 22783), subject to a data transfer agreement. Ethics approval for the UK Biobank study was obtained from the North West Centre for Research Ethics Committee (11/NW/0382). For the PNC study, the institutional review boards of both the University of Pennsylvania and the Children’s Hospital of Philadelphia approved all study procedures. Informed consent was obtained from all subjects. We gratefully acknowledge all the studies and databases that made their GWAS summary data available. The authors acknowledge the Texas Advanced Computing Center (TACC, http://www.tacc.utexas.edu) at The University of Texas at Austin for providing HPC and storage resources that have contributed to the research results reported within this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhao, B., Zhang, J., Ibrahim, J.G. et al. Large-scale GWAS reveals genetic architecture of brain white matter microstructure and genetic overlap with cognitive and mental health traits (n = 17,706). Mol Psychiatry 26, 3943–3955 (2021). https://doi.org/10.1038/s41380-019-0569-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-019-0569-z
This article is cited by
-
High-dimensional mediation analysis reveals the mediating role of physical activity patterns in genetic pathways leading to AD-like brain atrophy
BioData Mining (2025)
-
Whole exome sequencing identified six novel genes for depressive symptoms
Molecular Psychiatry (2025)
-
Genome-wide analysis identifies novel shared loci between depression and white matter microstructure
Molecular Psychiatry (2025)
-
Enhanced insights into the genetic architecture of 3D cranial vault shape using pleiotropy-informed GWAS
Communications Biology (2025)
-
Genetic Insights into Brain Morphology: a Genome-Wide Association Study of Cortical Thickness and T1-Weighted MRI Gray Matter-White Matter Intensity Contrast
Neuroinformatics (2025)