Abstract
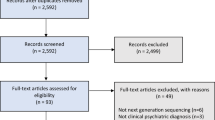
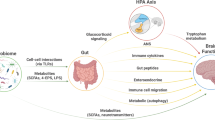
Psychiatric disorders pose substantial global burdens on public health, yet therapeutic options remain limited. Recently, gut microbiota is in the spotlight of new research on psychiatric disorders, as emerging discoveries have highlighted the importance of gut microbiome in the regulation of central nervous system via mediating the gut-brain-axis bidirectional communication. While metagenomics studies have accumulated for psychiatric disorders, few systematic efforts were dedicated to integrating these high-throughput data across diverse phenotypes, interventions, geographical regions, and biological species. To present a panoramic view of global data and provide a comprehensive resource for investigating the gut microbiota dysbiosis in psychiatric disorders, we developed the PsycGM, a manually curated and well-annotated database that provides the literature-supported associations between gut microbiota and psychiatric disorders or intervention measures. In total, PsycGM incorporated 559 studies from 31 countries worldwide, encompassing research involving humans, rats, mice, and non-human primates. PsycGM documented 8907 curated associations between 1514 gut microbial taxa and 11 psychiatric disorders, as well as 4050 associations between 869 taxa and 232 microbiota-based and non-microbiota-based interventions. Moreover, PsycGM provided a user-friendly web interface with comprehensive information, enabling browsing, retrieving and downloading of all entries. In the application of PsycGM, we panoramically depicted the intestinal microecological imbalance in depression. Additionally, we identified 9 microbial taxa consistently altered in patients with depression, with the most common dysregulations observed for Parabacteroides, Alistipes, and Faecalibacterium; in animal models of depression, consistent changes were observed in 21 microbial taxa, most frequently reported as Helicobacter, Lactobacillus, Roseburia, and the ratio of Firmicutes/Bacteroidetes. PsycGM is a comprehensive resource for future investigations on the role of gut microbiota in mental and brain health, and for therapeutic target innovations based on modifications of gut microbiota. PsycGM is freely accessed at http://psycgmomics.info.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
PsycGM is freely available at http://psycgmomics.info. All data sources analyzed in this study are included in the Supplementary Information. Supplementary information is available at MP’s website.
References
O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93.
Postler TS, Ghosh S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab. 2017;26:110–30.
Lee JY, Tsolis RM, Bäumler AJ. The microbiome and gut homeostasis. Science. 2022;377:eabp9960.
de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut. 2022;71:1020–32.
Cryan JF, O’Riordan KJ, Cowan CSM, Sandhu KV, Bastiaanssen TFS, Boehme M, et al. The microbiota-gut-brain axis. Physiol Rev. 2019;99:1877–2013.
Socała K, Doboszewska U, Szopa A, Serefko A, Włodarczyk M, Zielińska A, et al. The role of microbiota-gut-brain axis in neuropsychiatric and neurological disorders. Pharmacol Res. 2021;172:105840.
Loh JS, Mak WQ, Tan LKS, Ng CX, Chan HH, Yeow SH, et al. Microbiota-gut-brain axis and its therapeutic applications in neurodegenerative diseases. Signal Transduct Target Ther. 2024;9:37.
Liu L, Wang H, Zhang H, Chen X, Zhang Y, Wu J, et al. Toward a deeper understanding of gut microbiome in depression: the promise of clinical applicability. Adv Sci. 2022;9:e2203707.
Borkent J, Ioannou M, Laman JD, Haarman BCM, Sommer IEC. Role of the gut microbiome in three major psychiatric disorders. Psychol Med. 2022;52:1222–42.
Morais LH, Schreiber HL, Mazmanian SK. The gut microbiota-brain axis in behaviour and brain disorders. Nat Rev Microbiol. 2021;19:241–55.
Ma Q, Xing C, Long W, Wang HY, Liu Q, Wang RF. Impact of microbiota on central nervous system and neurological diseases: the gut-brain axis. J Neuroinflammation. 2019;16:53.
Vuotto C, Battistini L, Caltagirone C, Borsellino G. Gut microbiota and disorders of the central nervous system. Neuroscientist. 2020;26:487–502.
Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–79.
Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry. 2016;21:786–96.
Zheng P, Wu J, Zhang H, Perry SW, Yin B, Tan X, et al. The gut microbiome modulates gut-brain axis glycerophospholipid metabolism in a region-specific manner in a nonhuman primate model of depression. Mol Psychiatry. 2021;26:2380–92.
Zheng P, Zeng B, Liu M, Chen J, Pan J, Han Y, et al. The gut microbiome from patients with schizophrenia modulates the glutamate-glutamine-GABA cycle and schizophrenia-relevant behaviors in mice. Sci Adv. 2019;5:eaau8317.
Zheng P, Yang J, Li Y, Wu J, Liang W, Yin B, et al. Gut microbial signatures can discriminate unipolar from bipolar depression. Adv Sci. 2020;7:1902862.
Burokas A, Arboleya S, Moloney RD, Peterson VL, Murphy K, Clarke G, et al. Targeting the microbiota-gut-brain axis: prebiotics have anxiolytic and antidepressant-like effects and reverse the impact of chronic stress in mice. Biol Psychiatry. 2017;82:472–87.
Abildgaard A, Kern T, Pedersen O, Hansen T, Wegener G, Lund S. The antidepressant-like effect of probiotics and their faecal abundance may be modulated by the cohabiting gut microbiota in rats. Eur Neuropsychopharmacol. 2019;29:98–110.
Getachew B, Aubee JI, Schottenfeld RS, Csoka AB, Thompson KM, Tizabi Y. Ketamine interactions with gut-microbiota in rats: relevance to its antidepressant and anti-inflammatory properties. BMC Microbiol. 2018;18:222.
Hashimoto K. Emerging role of the host microbiome in neuropsychiatric disorders: overview and future directions. Mol Psychiatry. 2023;28:3625–37.
Kodama Y, Shumway M, Leinonen R. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–6.
Harrison PW, Alako B, Amid C, Cerdeño-Tárraga A, Cleland I, Holt S, et al. The European Nucleotide Archive in 2018. Nucleic Acids Res. 2019;47:D84–8.
Okido T, Kodama Y, Mashima J, Kosuge T, Fujisawa T, Ogasawara O. DNA Data Bank of Japan (DDBJ) update report 2021. Nucleic Acids Res. 2022;50:D102–5.
Cheng L, Qi C, Zhuang H, Fu T, Zhang X. gutMDisorder: a comprehensive database for dysbiosis of the gut microbiota in disorders and interventions. Nucleic Acids Res. 2020;48:D554–60.
Zhang Q, Yu K, Li S, Zhang X, Zhao Q, Zhao X, et al. gutMEGA: a database of the human gut MEtaGenome Atlas. Brief Bioinform. 2021;22:bbaa082.
Wu S, Sun C, Li Y, Wang T, Jia L, Lai S, et al. GMrepo: a database of curated and consistently annotated human gut metagenomes. Nucleic Acids Res. 2020;48:D545–53.
Ruszkowski J, Daca A, Szewczyk A, Dębska-Ślizień A, Witkowski JM. The influence of biologics on the microbiome in immune-mediated inflammatory diseases: a systematic review. Biomed Pharmacother. 2021;141:111904.
Bock PM, Martins AF, Ramalho R, Telo GH, Leivas G, Maraschin CK, et al. The impact of dietary, surgical, and pharmacological interventions on gut microbiota in individuals with diabetes mellitus: a systematic review. Diabetes Res Clin Pract. 2022;189:109944.
McGuinness AJ, Loughman A, Foster JA, Jacka F. Mood disorders: the gut bacteriome and beyond. Biol Psychiatry. 2024;95:319–28.
Jayaraman S. Of ethnicity, environment, and microbiota. Cell Mol Immunol. 2019;16:106–8.
Parizadeh M, Arrieta MC. The global human gut microbiome: genes, lifestyles, and diet. Trends Mol Med. 2023;29:789–801.
Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, et al. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci USA. 2014;111:E2329–38.
Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179–89.
Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–81.
Janssens Y, Nielandt J, Bronselaer A, Debunne N, Verbeke F, Wynendaele E, et al. Disbiome database: linking the microbiome to disease. BMC Microbiol. 2018;18:50.
Pu J, Yu Y, Liu Y, Tian L, Gui S, Zhong X, et al. MENDA: a comprehensive curated resource of metabolic characterization in depression. Brief Bioinform. 2020;21:1455–64.
Zhang L, Chang S, Li Z, Zhang K, Du Y, Ott J, et al. ADHDgene: a genetic database for attention deficit hyperactivity disorder. Nucleic Acids Res. 2012;40:D1003–009.
Cheng L, Yang H, Zhao H, Pei X, Shi H, Sun J, et al. MetSigDis: a manually curated resource for the metabolic signatures of diseases. Brief Bioinform. 2019;20:203–9.
Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, et al. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database. 2020;2020:baaa062.
Rikke BA, Wynes MW, Rozeboom LM, Barón AE, Hirsch FR. Independent validation test of the vote-counting strategy used to rank biomarkers from published studies. Biomark Med. 2015;9:751–61.
Park JE, Lim HR, Kim JW, Shin KH. Metabolite changes in risk of type 2 diabetes mellitus in cohort studies: a systematic review and meta-analysis. Diabetes Res Clin Pract. 2018;140:216–27.
Acknowledgements
This study was supported by the Joint Project of Chongqing Municipal Science and Technology Bureau and Chongqing Health Commission (2023CCXM003), the Natural Science Foundation of Chongqing (CSTB2024NSCQ-MSX1027), the Fund of Moutaintop Plan Project in the First Affiliated Hospital of Chongqing Medical University (cyyy-xkdfjh-lcyj-202301), the Natural Science Foundation Project of China (82371526), and the Natural Science Foundation Project of Chongqing (cstc2022ycjh-bgzxm0033). We thank Zhixin Chen for optimizing and beautifying the webpage of this database.
Author information
Authors and Affiliations
Contributions
Xie P and Liu YY conceptualized and designed the study. Wang DF was responsible for the literature search. Wang DF, Gui SW, Pu JC, Yan L, Li ZC, Tao XK were responsible for the study selection. Wang DF, Gui SW, Pu JC, Zhong XG, Yan L, Li ZC, Tao XK, Yang D, Zhou HP, and Qiao RJ were responsible for the data extraction, data curation, and quality assessment. Wang DF, Gui SW, and Pu JC wrote the original draft. Zhang HP, Cheng XY, Ren Y, Chen WY, Chen XP, Tao W, Chen Y, Chen X, Liu YY, and Xie P commented on and revised the manuscript. Xie P and Liu YY reviewed, edited, and made the final version. All authors contributed to the final draft of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations. This study was approved by The Ethics Committee of Chongqing Medical University (IACUC-CQMU-2024-0115). The datasets generated in the current study were collected from publicly available literature or reports, therefore informed consent forms are not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, D., Gui, S., Pu, J. et al. PsycGM: a comprehensive database for associations between gut microbiota and psychiatric disorders. Mol Psychiatry (2025). https://doi.org/10.1038/s41380-025-03000-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-025-03000-5