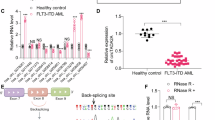
Abstract
AML is a complex disease caused by multiple molecular mechanisms. As an important regulatory molecule, the role of circRNA in AML is not fully understood. By performing high-throughput sequencing on clinical samples, we systematically identified the differences in circRNA expression and distribution between AML and healthy donor samples. One circular RNA, circAFF2, was found to be significantly upregulated in AML patients. Functional studies showed that knockdown of circAFF2 could significantly inhibit the proliferation of AML cells and promote their apoptosis. Overexpression of circAFF2 can have opposite effects. In vivo experiments showed that transplantation of AML cells with circAFF2 knockdown slowed the proliferation and infiltration and prolonged the survival time of mice compared to controls. Further studies showed that circAFF2 can promote the degradation of PML mRNA by binding to the 3’UTR of PML mRNA, thereby affecting the proliferation and apoptosis of AML cells. In conclusion, our work demonstrates that circAFF2 can bind to PML mRNA to regulate AML cell function, providing new insights into the mechanism of AML development and potential targets for clinical diagnosis and treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
269,00 € per year
only 5,38 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All raw data can be accessed in the NCBI GEO database (GSE229582).
References
Acute myeloid leukaemia. Nat Rev Dis Prim. 2016 2:16011.
The 49th Annual Meeting of the European Society for Blood and Marrow Transplantation: Physicians – Poster Session (P001-P708). Bone Marrow Transplant. 2023; 58:153–627.
Bhatt VR, Uy GL, Klepin HD. Determining treatment tolerance and fitness for intensive chemotherapy in older adults with AML: a call to action. Blood. 2024;143:483–7.
Harbi S, Brac de la Perriere L, Bouchacourt B, Garciaz S, Pagliardini T, Calmels B, et al. Peripheral blood haploidentical hematopoietic cell transplantation for patients aged 70 years and over with acute myeloid leukemia or high-risk myelodysplastic syndrome. Bone Marrow Transpl. 2024;59:101–6.
Bose P, Konopleva MY. ORY-1001: overcoming the differentiation block in AML. Cancer Cell. 2018;33:342–3.
Philip SJ, Visconte V, Carraway HE, Gerds AT, Singh A, Balderman S, et al. Significance of the apoptotic regulators BAX, BCL2, BCL- XL and MCL1 in newly diagnosed AML. Blood. 2023;142:5995–5995.
Shao X, Xu A, Du W, Xu T, Huang Y, Xia Z, et al. The palmitoyltransferase ZDHHC21 regulates oxidative phosphorylation to induce differentiation block and stemness in AML. Blood. 2023;142:365–81.
Chen B, Huang S. Circular RNA: An emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41–50.
Ma X, Ying Y, Sun J, Xie H, Li J, He L, et al. circKDM4C enhances bladder cancer invasion and metastasis through miR-200bc-3p/ZEB1 axis. Cell Death Discov. 2021;7:365.
Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–9.
Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–83.
Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016;143:1838–47.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91.
Sun YM, Wang WT, Zeng ZC, Chen TQ, Han C, Pan Q, et al. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood. 2019;134:1533–46.
Yang X, Liu J, Liu W, Wu H, Wei Y, Guo X, et al. circFAM193B interaction with PRMT6 regulates AML leukemia stem cells chemoresistance through altering the oxidative metabolism and lipid peroxidation. Leukemia. 2024.
Wei W, Pan J, Wang J, Mao S, Qian Y, Lin X, et al. circSLC25A13 acts as a ceRNA to regulate AML progression via miR-616-3p/ADCY2 axis. Mol Carcinog. 2023;62:1546–62.
Li K, Guo J, Ming Y, Chen S, Zhang T, Ma H, et al. A circular RNA activated by TGFβ promotes tumor metastasis through enhancing IGF2BP3-mediated PDPN mRNA stability. Nat Commun. 2023;14:6876.
Rossi F, Beltran M, Damizia M, Grelloni C, Colantoni A, Setti A, et al. Circular RNA ZNF609/CKAP5 mRNA interaction regulates microtubule dynamics and tumorigenicity. Mol cell. 2022;82:75–89.e79.
Wu S, Xu H, Zhang R, Wang X, Yang J, Li X, et al. Circular RNA circLAMA3 inhibits the proliferation of bladder cancer by directly binding an mRNA. Mol Ther Oncolytics. 2022;24:742–54.
Yu X, Tong H, Chen J, Tang C, Wang S, Si Y, et al. CircRNA MBOAT2 promotes intrahepatic cholangiocarcinoma progression and lipid metabolism reprogramming by stabilizing PTBP1 to facilitate FASN mRNA cytoplasmic export. Cell Death Dis. 2023;14:20.
Ji Z, Lu R, Wu T, Chen Z, Wen Z, Li Z, et al. Expression profiling of circular RNA reveals a potential miR-145-5p sponge function of circ-AFF2 and circ-ASAP1 in renal cell carcinoma. Am J Transl Res. 2023;15:82–98.
Shao Y, Liu Z, Song X, Sun R, Zhou Y, Zhang D, et al. ALKBH5/YTHDF2-mediated m6A modification of circAFF2 enhances radiosensitivity of colorectal cancer by inhibiting Cullin neddylation. Clin Transl Med. 2023;13:e1318.
Zhou F, Wang D, Wei W, Chen H, Shi H, Zhou N, et al. Comprehensive profiling of circular RNA expressions reveals potential diagnostic and prognostic biomarkers in multiple myeloma. BMC Cancer. 2020;20:40.
Lux S, Blatte TJ, Gillissen B, Richter A, Cocciardi S, Skambraks S, et al. Deregulated expression of circular RNAs in acute myeloid leukemia. Blood Adv. 2021;5:1490–503.
Whiteley AE, Price TT, Cantelli G, Sipkins DA. Leukaemia: a model metastatic disease. Nat Rev Cancer. 2021;21:461–75.
Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:165–70.
Mann M, Wright PR, Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45:W435–W439.
Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–65.
Pisignano G, Michael DC, Visal TH, Pirlog R, Ladomery M, Calin GA. Going circular: history, present, and future of circRNAs in cancer. Oncogene. 2023;42:2783–2800.
Singh V, Uddin MH, Zonder JA, Azmi AS, Balasubramanian SK. Circular RNAs in acute myeloid leukemia. Mol Cancer. 2021;20:149.
Nair R, Salinas-Illarena A, Baldauf HM. New strategies to treat AML: novel insights into AML survival pathways and combination therapies. Leukemia. 2021;35:299–311.
Sawyers CL, Denny CT, Witte ON. Leukemia and the disruption of normal hematopoiesis. Cell. 1991;64:337–50.
Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101.
Bernardi R, Guernah I, Jin D, Grisendi S, Alimonti A, Teruya-Feldstein J, et al. PML inhibits HIF-1α translation and neoangiogenesis through repression of mTOR. Nature. 2006;442:779–85.
Daniels MJ, Marson A, Venkitaraman AR. PML bodies control the nuclear dynamics and function of the CHFR mitotic checkpoint protein. Nat Struct Mol Biol 2004;11:1114–21.
Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W, et al. The function of PML in p53-dependent apoptosis. Nat Cell Biol. 2000;2:730–6.
Li L, He D, He H, Wang X, Zhang L, Luo Y, et al. Overexpression of PML induced apoptosis in bladder cancer cell by caspase-dependent pathway. Cancer Lett. 2006;236:259–68.
Singh N, Sobti RC, Suri V, Nijhawan R, Sharma S, Das BC, et al. Downregulation of tumor suppressor gene PML in uterine cervical carcinogenesis: impact of human papillomavirus infection (HPV). Gynecol Oncol. 2013;128:420–6.
Vancurova M, Hanzlikova H, Knoblochova L, Kosla J, Majera D, Mistrik M, et al. PML nuclear bodies are recruited to persistent DNA damage lesions in an RNF168-53BP1-dependent manner and contribute to DNA repair. DNA Repair. 2019;78:114–27.
Li X, Ding J, Wang X, Cheng Z, Zhu Q. NUDT21 regulates circRNA cyclization and ceRNA crosstalk in hepatocellular carcinoma. Oncogene. 2020;39:891–904.
Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun. 2016;7:12060.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.
Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–80.
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–87.
Feng J, Chen W, Dong X, Wang J, Mei X, Deng J, et al. CSCD2: an integrated interactional database of cancer-specific circular RNAs. Nucleic Acids Res. 2022;50:D1179–D1183.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2018;7:1–6.
Dobin A, Gingeras TR. Mapping RNA-seq reads with STAR. Curr Protoc Bioinforma. 2015;51:11 14 11–11 14 19.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Xu L, Lyu M, Yang S, Zhang J, Yu D. CircRNA expression profiles of breast cancer and construction of a circRNA-miRNA-mRNA network. Sci Rep. 2022;12:17765.
Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98.
Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4.
Lai WS, Arvola RM, Goldstrohm AC, Blackshear PJ. Inhibiting transcription in cultured metazoan cells with actinomycin D to monitor mRNA turnover. Methods. 2019;155:77–87.
Lv K, Ren JG, Han X, Gui J, Gong C, Tong W. Depalmitoylation rewires FLT3-ITD signaling and exacerbates leukemia progression. Blood. 2021;138:2244–55.
Cardiff RD, Miller CH, Munn RJ. Manual hematoxylin and eosin staining of mouse tissue sections. Cold Spring Harb Protoc. 2014;2014:655–8.
Southern EM. Blotting at 25. Trends Biochem Sci. 2000;25:585–8.
Tan S, Kang Y, Li H, He HQ, Zheng L, Wu SQ, et al. circST6GALNAC6 suppresses bladder cancer metastasis by sponging miR-200a-3p to modulate the STMN1/EMT axis. Cell Death Dis. 2021;12:168.
Huang G, Liang M, Liu H, Huang J, Li P, Wang C, et al. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis. 2020;11:1065.
Funding
This research was funded by the National Natural Science Foundation of China (82170170, 81870129, and 82173314) and the Fundamental Research Funds for the Central Universities (2662022SYYJ001).
Author information
Authors and Affiliations
Contributions
CH, SC, and KC conceived, designed, and supervised the project. HL, LL, and MF provided technical assistance. LY, XZ, XL, JX, SY, FL, YS, LR, LC, and WY collected samples and performed experiments. WC, CZ, and SL performed the bioinformatics analysis. CH, LY, SC, and KC drafted the manuscript with contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and informed consent
The study was approved by the Ethics Committee (Institutional Review Board) of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology (Approved No. of ethics committee: Lingbo Liu 2023#0508). All patients provided written informed consent for participation according to the Declaration of Helsinki and written informed consent for publication. Clinical information was obtained from the medical records. The animal experiments in this study were approved by the Animal Experimental Ethical Committee of the Huazhong Agricultural University. All in vivo operations were performed in accordance with the Guidelines for Animal Experimentation of Huazhong Agricultural University and the National Institute of Health Guide for the Care and Use of Laboratory Animals.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yao, L., Zhang, X., Li, X. et al. circAFF2 promotes the development of AML by binding to PML mRNA. Oncogene 44, 1234–1244 (2025). https://doi.org/10.1038/s41388-025-03299-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-025-03299-y