Abstract
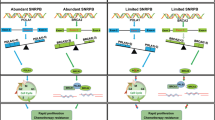
Dysfunction or aberrant expression of DEAD-box RNA helicases might play a role in the initiation and progression of human cancers. Nevertheless, the key regulator and underlying molecular mechanism have yet to be fully elucidated in ovarian cancer. This study identified DDX39A as one of the prominently upregulated genes in ovarian cancer through a systematic analysis of RNA helicase expression profiles using the CPTAC and TCGA ovarian cancer datasets. High expression of DDX39A was confirmed in paraffin-embedded ovarian cancer samples. Specifically, elevated DDX39A expression was found to be associated with poor overall survival in ovarian cancer patients. Antisense oligonucleotide-mediated DDX39A silencing led to a decrease in the proliferation capacity of a CDX model and a PDX model. Furthermore, DDX39A expression is regulated by the splicing factor SNRPB. SNRPB depletion or DDX39A knockdown induced the retention of DDX39A introns 6 and 8 to generate the noncoding transcript DDX39A-209, which yielded premature termination codons and resulted in nonsense-mediated RNA decay and decreased expression of the DDX39A protein. DDX39A silencing reduced the proliferative and metastatic capacities of SNRPB-overexpressing cells, indicating that DDX39A mediates the oncogenic function of SNRPB in ovarian cancer cells. In addition, RNA-Seq data analysis revealed that DDX39A promotes the proliferation and metastasis of ovarian cancer cells through the regulation of exon skipping of ITGA6 to produce the oncogenic ITGA6A transcript. These findings suggest that the SNRPB/DDX39A/ITGA6 axis plays critically important role in the progression of ovarian cancer, which increases our understanding of the role of DEAD-box RNA helicases and provides a viable therapeutic target for ovarian cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
269,00 € per year
only 5,38 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48.
Cotto KC, Feng YY, Ramu A, Richters M, Freshour SL, Skidmore ZL, et al. Integrated analysis of genomic and transcriptomic data for the discovery of splice-associated variants in cancer. Nat Commun. 2023;14:1589.
Zhang Y, Qian J, Gu C, Yang Y. Alternative splicing and cancer: a systematic review. Signal Transduct Target Ther. 2021;6:78.
Bradley RK, Anczukow O. RNA splicing dysregulation and the hallmarks of cancer. Nat Rev Cancer. 2023;23:135–55.
Linder P, Jankowsky E. From unwinding to clamping—the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–16.
Ali MAM. The DEAD-box protein family of RNA helicases: sentinels for a myriad of cellular functions with emerging roles in tumorigenesis. Int J Clin Oncol. 2021;26:795–825.
Zhou HZ, Li F, Cheng ST, Xu Y, Deng HJ, Gu DY, et al. DDX17-regulated alternative splicing that produced an oncogenic isoform of PXN-AS1 to promote HCC metastasis. Hepatology. 2022;75:847–65.
Zhao G, Yuan H, Li Q, Zhang J, Guo Y, Feng T, et al. DDX39B drives colorectal cancer progression by promoting the stability and nuclear translocation of PKM2. Signal Transduct Target Ther. 2022;7:275.
Le TK, Cherif C, Omabe K, Paris C, Lannes F, Audebert S, et al. DDX5 mRNA-targeting antisense oligonucleotide as a new promising therapeutic in combating castration-resistant prostate cancer. Mol Ther. 2023;31:471–86.
Sugiura T, Nagano Y, Noguchi Y. DDX39, upregulated in lung squamous cell cancer, displays RNA helicase activities and promotes cancer cell growth. Cancer Biol Ther. 2007;6:957–64.
Wang X, Li P, Wang C, Zhang D, Zeng L, Liu X, et al. DEAD-box RNA Helicase 39 Promotes Invasiveness and Chemoresistance of ER-positive Breast Cancer. J Cancer. 2020;11:1846–58.
Bao Y, Jiang A, Dong K, Gan X, Gong W, Wu Z, et al. DDX39 as a predictor of clinical prognosis and immune checkpoint therapy efficacy in patients with clear cell renal cell carcinoma. Int J Biol Sci. 2021;17:3158–72.
Diao Y, Li Y, Wang Z, Wang S, Li P, Kong B. SF3B4 promotes ovarian cancer progression by regulating alternative splicing of RAD52. Cell Death Dis. 2022;13:179.
Li Y, Chen Z, Peng J, Yuan C, Yan S, Yang N, et al. The splicing factor SNRPB promotes ovarian cancer progression through regulating aberrant exon skipping of POLA1 and BRCA2. Oncogene. 2023;42:2386–401.
Goel HL, Gritsko T, Pursell B, Chang C, Shultz LD, Greiner DL, et al. Regulated splicing of the alpha6 integrin cytoplasmic ___domain determines the fate of breast cancer stem cells. Cell Rep. 2014;7:747–61.
Li F, Zhao C, Diao Y, Wang Z, Peng J, Yang N, et al. MEX3A promotes the malignant progression of ovarian cancer by regulating intron retention in TIMELESS. Cell Death Dis. 2022;13:553.
Wei Y, Chen Z, Li Y, Song K. The splicing factor WBP11 mediates MCM7 intron retention to promote the malignant progression of ovarian cancer. Oncogene. 2024;43:1565–78.
Edwards NJ, Oberti M, Thangudu RR, Cai S, McGarvey PB, Jacob S, et al. The CPTAC data portal: a resource for cancer proteomics research. J Proteome Res. 2015;14:2707–13.
Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–8.
Gyorffy B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience. 2023;45:1889–98.
Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560.
de Bruijn I, Kundra R, Mastrogiacomo B, Tran TN, Sikina L, Mazor T, et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023;83:3861–7.
Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216–W221.
Tan TZ, Yang H, Ye J, Low J, Choolani M, Tan DS, et al. CSIOVDB: a microarray gene expression database of epithelial ovarian cancer subtype. Oncotarget. 2015;6:43843–52.
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant. 2020;13:1194–202.
Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111:E5593–601.
Integrated genomic analyses of ovarian carcinoma. 2011;474:609-15. pCancer Genome AtlasResearch, N.
Groulx JF, Boudjadi S, and Beaulieu JF. MYC regulates alpha6 integrin subunit expression and splicing under its pro-proliferative ITGA6A form in colorectal cancer cells. Cancers. 2018;10.
Wang Y, Li L, Zhang X, Zhao X. Long non-coding RNA OIP5-AS1 suppresses microRNA-92a to augment proliferation and metastasis of ovarian cancer cells through upregulating ITGA6. J Ovarian Res. 2022;15:25.
Wei L, Yin F, Chen C, Li L. Expression of integrin alpha-6 is associated with multi drug resistance and prognosis in ovarian cancer. Oncol Lett. 2019;17:3974–80.
Bohnsack KE, Yi S, Venus S, Jankowsky E, Bohnsack MT. Cellular functions of eukaryotic RNA helicases and their links to human diseases. Nat Rev Mol Cell Biol. 2023;24:749–69.
Naineni SK, Robert F, Nagar B, Pelletier J. Targeting DEAD-box RNA helicases: The emergence of molecular staples. Wiley Interdiscip Rev RNA. 2023;14:e1738.
Andrisani O, Liu Q, Kehn P, Leitner WW, Moon K, Vazquez-Maldonado N, et al. Biological functions of DEAD/DEAH-box RNA helicases in health and disease. Nat Immunol. 2022;23:354–7.
Zhang T, Ma Z, Liu L, Sun J, Tang H, Zhang B, et al. DDX39 promotes hepatocellular carcinoma growth and metastasis through activating Wnt/beta-catenin pathway. Cell Death Dis. 2018;9:675.
Kuramitsu Y, Suenaga S, Wang Y, Tokuda K, Kitagawa T, Tanaka T, et al. Up-regulation of DDX39 in human pancreatic cancer cells with acquired gemcitabine resistance compared to gemcitabine-sensitive parental cells. Anticancer Res. 2013;33:3133–6.
Liu N, Wu Z, Chen A, Wang Y, Cai D, Zheng J, et al. SNRPB promotes the tumorigenic potential of NSCLC in part by regulating RAB26. Cell Death Dis. 2019;10:667.
Bourgeois CF, Mortreux F, Auboeuf D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat Rev Mol Cell Biol. 2016;17:426–38.
Kouyama Y, Masuda T, Fujii A, Ogawa Y, Sato K, Tobo T, et al. Oncogenic splicing abnormalities induced by DEAD-Box Helicase 56 amplification in colorectal cancer. Cancer Sci. 2019;110:3132–44.
Lu CA, Huang CK, Huang WS, Huang TS, Liu HY, Chen YF. DEAD-Box RNA helicase 42 plays a critical role in pre-mRNA splicing under cold stress. Plant Physiol. 2020;182:255–71.
Hirano M, Galarza-Munoz G, Nagasawa C, Schott G, Wang L, Antonia AL, et al. The RNA helicase DDX39B activates FOXP3 RNA splicing to control T regulatory cell fate. Elife. 2023;12.
Brooks DL, Schwab LP, 26 Krutilina R, Parke DN, Sethuraman A, Hoogewijs D, et al. ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models. Mol Cancer. 2016;15:26.
Ammothumkandy A, Maliekal TT, Bose MV, Rajkumar T, Shirley S, Thejaswini B, et al. CD66 and CD49f expressing cells are associated with distinct neoplastic phenotypes and progression in human cervical cancer. Eur J Cancer. 2016;60:166–78.
Jin H, Ying X, Que B, Wang X, Chao Y, Zhang H, et al. N(6)-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019;47:195–207.
Gambelli A, Nespolo A, Rampioni Vinciguerra GL, Pivetta E, Pellarin I, Nicoloso MS, et al. Platinum-induced upregulation of ITGA6 promotes chemoresistance and spreading in ovarian cancer. EMBO Mol Med. 2024;16:1162–92.
Asada T, Nakahata S, Fauzi YR, Ichikawa T, Inoue K, Shibata N, et al. Integrin alpha6A (ITGA6A)-type splice variant in extracellular vesicles has a potential as a novel marker of the early recurrence of pancreatic cancer. Anticancer Res. 2022;42:1763–75.
Acknowledgements
We thank American Journal Experts (AJE) for English language editing.
Funding
This work was supported by Natural Science Foundation of Shandong Province (ZR2023MH183, ZR2022QH074), the Tai-Shan Scholar Program of Shandong Province (No. ts20070743).
Author information
Authors and Affiliations
Contributions
Conception and design: YW L. Methodology: YW L. Acquisition of data: YW L, ZC, YL. Analysis and interpretation of data: YW L, ZC, YL. Administrative, technical, or material support: YW L, ZC, YL, YG, HX, QG, YP, LG, YD, CY, SY, NY. Study supervision: YW L, BK. Writing, review, and/or revision of the manuscript: YW L Final approval: All authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All methods in this study were performed in accordance with the relevant guidelines and regulations. Ethics Committee at Qilu Hospital of Shandong University approved the study (KYLL-202407-059-1). The Nude mouse xenograft assay and PDX model assay were approved by the Shandong University Animal Care and Use Committee (24045). Informed consent was obtained from all participants.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Chen, Z., Gao, Y. et al. SNRPB-mediated regulation of DDX39A splicing promotes ovarian cancer progression by regulating α6 integrin subunit expression. Oncogene 44, 2170–2185 (2025). https://doi.org/10.1038/s41388-025-03386-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-025-03386-0