Abstract
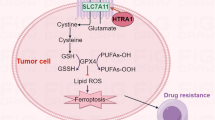
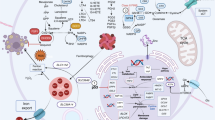
Ferroptosis is a newly discovered type of regulated cell death, characterized by the iron-dependent accumulation of lipid reactive oxygen species, which has been implicated in a number of human diseases. However, the regulatory mechanisms underlying ferroptosis in colorectal cancer (CRC) remain unclear. In this study, we unravel the pivotal role of PRMT5 in the progression of CRC by promoting ferroptosis resistance. Mechanistically, PRMT5 directly inhibits the transcription of m6A demethylase ALKBH5 via histone modifications (H4R3me2s and H3R8me2s), bolstering SLC7A11 mRNA stability and expression, thereby aggravating CRC progression through attenuating ferroptosis. Particularly, our work identifies PRMT5 as a novel lactylation substrate at lysine 240 (PRMT5 K240lac), crucial for sustaining CRC ferroptosis resistance by shaping the ALKBH5/SLC7A11 axis, while mutation disrupting these effects. Overall, our work underscores the significance of PRMT5 K240lac in conferring ferroptosis resistance in CRC, proposing targeted intervention along the PRMT5 K240lac/ALKBH5/SLC7A11 axis as an innovative therapeutic approach in CRC treatment.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
269,00 € per year
only 5,38 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The datasets used in the current study are available from the corresponding author on reasonable request.
References
Yang WS, Stockwell BR. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26:165–76.
Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22:266–82.
Duan JY, Lin X, Xu F, Shan SK, Guo B, Li FX, et al. Ferroptosis and its potential role in metabolic diseases: a curse or revitalization?. Front Cell Dev Biol. 2021;9:701788.
Du L, Wu Y, Fan Z, Li Y, Guo X, Fang Z, et al. The role of ferroptosis in nervous system disorders. J Integr Neurosci. 2023;22:19.
Fang X, Ardehali H, Min J, Wang F. The molecular and metabolic landscape of iron and ferroptosis in cardiovascular disease. Nat Rev Cardiol. 2023;20:7–23.
Ni L, Yuan C, Wu X. Targeting ferroptosis in acute kidney injury. Cell Death Dis. 2022;13:182.
Geng Z, Guo Z, Guo R, Ye R, Zhu W, Yan B. Ferroptosis and traumatic brain injury. Brain Res Bull. 2021;172:212–19.
Sun Y, Chen P, Zhai B, Zhang M, Xiang Y, Fang J, et al. The emerging role of ferroptosis in inflammation. Biomed Pharmacother. 2020;127:110108.
Ouyang S, You J, Zhi C, Li P, Lin X, Tan X, et al. Ferroptosis: the potential value target in atherosclerosis. Cell Death Dis. 2021;12:782.
Du G, Zhang Q, Huang X, Wang Y. Molecular mechanism of ferroptosis and its role in the occurrence and treatment of diabetes. Front Genet. 2022;13:1018829.
Wang Y, Wei Z, Pan K, Li J, Chen Q. The function and mechanism of ferroptosis in cancer. Apoptosis. 2020;25:786–98.
Lei G, Zhuang L, Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer. 2022;22:381–96.
Liang C, Zhang X, Yang M, Dong X. Recent progress in ferroptosis inducers for cancer therapy. Adv Mater. 2019;31:e1904197.
Smith LM, Kelleher NL. Consortium for Top Down P. Proteoform: a single term describing protein complexity. Nat Methods. 2013;10:186–7.
Nussinov R, Tsai CJ, Xin F, Radivojac P. Allosteric post-translational modification codes. Trends Biochem Sci. 2012;37:447–55.
Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol. 2006;16:288–302.
Li Z, Xu X. Post-translational modifications of the mini-chromosome maintenance proteins in DNA replication. Genes. 2019;10:331.
Chatterjee S, Senapati P, Kundu TK. Post-translational modifications of lysine in DNA-damage repair. Essays Biochem. 2012;52:93–111.
Lee JM, Hammaren HM, Savitski MM, Baek SH. Control of protein stability by post-translational modifications. Nat Commun. 2023;14:201.
Sales-Gil R, Vagnarelli P. How HP1 post-translational modifications regulate heterochromatin formation and maintenance. Cells. 2020;9:1460.
Kretova M, Selicky T, Cipakova I, Cipak L. Regulation of pre-mRNA splicing: indispensable role of post-translational modifications of splicing factors. Life (Basel). 2023;13:604.
Cuijpers SAG, Vertegaal ACO. Guiding mitotic progression by crosstalk between post-translational modifications. Trends Biochem Sci. 2018;43:251–68.
Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65:8–24.
Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem Sci. 2011;36:633–41.
Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci. 2015;72:2041–59.
Kim H, Ronai ZA. PRMT5 function and targeting in cancer. Cell Stress. 2020;4:199–215.
Abumustafa W, Zamer BA, Khalil BA, Hamad M, Maghazachi AA, Muhammad JS. Protein arginine N-methyltransferase 5 in colorectal carcinoma: Insights into mechanisms of pathogenesis and therapeutic strategies. Biomed Pharmacother. 2022;145:112368.
Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574:575–80.
Xie Y, Hu H, Liu M, Zhou T, Cheng X, Huang W, et al. The role and mechanism of histone lactylation in health and diseases. Front Genet. 2022;13:949252.
Liu X, Zhang Y, Li W, Zhou X. Lactylation, an emerging hallmark of metabolic reprogramming: Current progress and open challenges. Front Cell Dev Biol. 2022;10:972020.
Wang X, Fan W, Li N, Ma Y, Yao M, Wang G, et al. YY1 lactylation in microglia promotes angiogenesis through transcription activation-mediated upregulation of FGF2. Genome Biol. 2023;24:87.
Motolani A, Martin M, Sun M, Lu T. The structure and functions of PRMT5 in human diseases. Life (Basel). 2021;11:1074.
Yan H, Talty R, Johnson CH. Targeting ferroptosis to treat colorectal cancer. Trends Cell Biol. 2023;33:185–88.
Murakami S, Jaffrey SR. Hidden codes in mRNA: control of gene expression by m(6)A. Mol Cell. 2022;82:2236–51.
Qiu L, Jing Q, Li Y, Han J. RNA modification: mechanisms and therapeutic targets. Mol Biomed. 2023;4:25.
Xin Q, Wang H, Li Q, Liu S, Qu K, Liu C, et al. Lactylation: a passing fad or the future of posttranslational modification. Inflammation. 2022;45:1419–29.
Liu M, Yao B, Gui T, Guo C, Wu X, Li J, et al. PRMT5-dependent transcriptional repression of c-Myc target genes promotes gastric cancer progression. Theranostics. 2020;10:4437–52.
Wang Z, Li R, Hou N, Zhang J, Wang T, Fan P, et al. PRMT5 reduces immunotherapy efficacy in triple-negative breast cancer by methylating KEAP1 and inhibiting ferroptosis. J Immunother Cancer. 2023;11:e006890.
Fan Y, Wang Y, Dan W, Zhang Y, Nie L, Ma Z, et al. PRMT5-mediated arginine methylation stabilizes GPX4 to suppress ferroptosis in cancer. Nat Cell Biol. 2025;27:641–53.
Wu Y, Wang Z, Han L, Guo Z, Yan B, Guo L, et al. PRMT5 regulates RNA m6A demethylation for doxorubicin sensitivity in breast cancer. Mol Ther. 2022;30:2603–17.
Meng S, Liu H, Xu J, Deng C, Qian X, Chu S, et al. PRMT5-mediated ALKBH5 methylation promotes colorectal cancer immune evasion via increasing CD276 expression. Research (Wash D C). 2025;8:0549.
Qu J, Yan H, Hou Y, Cao W, Liu Y, Zhang E, et al. RNA demethylase ALKBH5 in cancer: from mechanisms to therapeutic potential. J Hematol Oncol. 2022;15:8.
Zhai J, Chen H, Wong CC, Peng Y, Gou H, Zhang J, et al. ALKBH5 drives immune suppression via targeting AXIN2 to promote colorectal cancer and is a target for boosting immunotherapy. Gastroenterology. 2023;165:445–62.
Shen D, Lin J, Xie Y, Zhuang Z, Xu G, Peng S, et al. RNA demethylase ALKBH5 promotes colorectal cancer progression by posttranscriptional activation of RAB5A in an m6A-YTHDF2-dependent manner. Clin Transl Med. 2023;13:e1279.
Luo J, Yu H, Yuan Z, Ye T, Hu B. ALKBH5 decreases SLC7A11 expression by erasing m6A modification and promotes the ferroptosis of colorectal cancer cells. Clin Transl Oncol. 2023;25:2265–76.
Huang Z, Lin G, Hong Y, Weng L, Zhu K, Zhuang W. High expression of AlkB homolog 5 suppresses the progression of non-small cell lung cancer by facilitating ferroptosis through m6A demethylation of SLC7A11. Environ Toxicol. 2024;39:4035–46.
Yang P, Wang Q, Liu A, Zhu J, Feng J. ALKBH5 holds prognostic values and inhibits the metastasis of colon cancer. Pathol Oncol Res. 2020;26:1615–23.
Ge J, Liu SL, Zheng JX, Shi Y, Shao Y, Duan YJ, et al. RNA demethylase ALKBH5 suppresses tumorigenesis via inhibiting proliferation and invasion and promoting CD8(+) T cell infiltration in colorectal cancer. Transl Oncol. 2023;34:101683.
Zhang Z, Wang L, Zhao L, Wang Q, Yang C, Zhang M, et al. N6-methyladenosine demethylase ALKBH5 suppresses colorectal cancer progression potentially by decreasing PHF20 mRNA methylation. Clin Transl Med. 2022;12:e940.
Wu X, Dai M, Li J, Cai J, Zuo Z, Ni S, et al. m(6)A demethylase ALKBH5 inhibits cell proliferation and the metastasis of colorectal cancer by regulating the FOXO3/miR-21/SPRY2 axis. Am J Transl Res. 2021;13:11209–22.
Pan RY, He L, Zhang J, Liu X, Liao Y, Gao J, et al. Positive feedback regulation of microglial glucose metabolism by histone H4 lysine 12 lactylation in Alzheimer’s disease. Cell Metab. 2022;34:634–48.e6.
Yu J, Chai P, Xie M, Ge S, Ruan J, Fan X, et al. Histone lactylation drives oncogenesis by facilitating m(6)A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 2021;22:85.
Lv X, Lv Y, Dai X. Lactate, histone lactylation and cancer hallmarks. Expert Rev Mol Med. 2023;25:e7.
Sun L, Zhang Y, Yang B, Sun S, Zhang P, Luo Z, et al. Lactylation of METTL16 promotes cuproptosis via m(6)A-modification on FDX1 mRNA in gastric cancer. Nat Commun. 2023;14:6523.
Chen Y, Wu J, Zhai L, Zhang T, Yin H, Gao H, et al. Metabolic regulation of homologous recombination repair by MRE11 lactylation. Cell. 2024;187:294–311.e21.
Xie B, Zhang M, Li J, Cui J, Zhang P, Liu F, et al. KAT8-catalyzed lactylation promotes eEF1A2-mediated protein synthesis and colorectal carcinogenesis. Proc Natl Acad Sci USA. 2024;121:e2314128121.
Cheng Z, Huang H, Li M, Chen Y. Proteomic analysis identifies PFKP lactylation in SW480 colon cancer cells. iScience. 2024;27:108645.
Zhong X, He X, Wang Y, Hu Z, Huang H, Zhao S, et al. Warburg effect in colorectal cancer: the emerging roles in tumor microenvironment and therapeutic implications. J Hematol Oncol. 2022;15:160.
Li H, Sun L, Gao P, Hu H. Lactylation in cancer: current understanding and challenges. Cancer Cell. 2024;42:1803–07.
Shen Y, Zhao P, Dong K, Wang J, Li H, Li M, et al. Tadalafil increases the antitumor activity of 5-FU through inhibiting PRMT5-mediated glycolysis and cell proliferation in colorectal cancer. Cancer Metab. 2022;10:22.
Funding
This work was supported by the Talent Construction Fund Research Project of Jiangsu Province Geriatric Hospital (Grant No. IR2024103), the Research Incubation Startup Fund of Jiangsu Province Geriatric Hospital (Grant No. FHQD202502), the Natural Science Foundation of Jiangsu Province (Grant No. BK202402101, BK20231261, BK20241997, BK20241998), the Jiangsu Primary Research & Development Plan (Grant No. BE2021747) and the Medical scientific research project of Jiangsu Provincial Commission of Health (Grant No. H2023048).
Author information
Authors and Affiliations
Contributions
ML, QD, LY, and SG provided the study concept and design. BY, ZY, and MX wrote the manuscript. SQ, BF, MX, YQ, LM, YJ, FH, JZ, MX, and JH performed the experiments. QZ and XZ interpreted and analyzed the data. YW and WX collected the patients’ samples. YJ, SG, BY, and ML provided financial support. ML, QD, LY, and SG is responsible for the overall content as the guarantor and supervised the project. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Written informed consent was obtained from all patients and the study was approved by the Ethics Committee of Jiangsu Province Geriatric Hospital. Animal experiments were performed according to the Health Guide for the Care and Use of Laboratory Animals approved by the Animal Experimental Research Ethics Committee of Jiangsu Province Geriatric Hospital.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Qu, S., Feng, B., Xing, M. et al. PRMT5 K240lac confers ferroptosis resistance via ALKBH5/SLC7A11 axis in colorectal cancer. Oncogene (2025). https://doi.org/10.1038/s41388-025-03457-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41388-025-03457-2