Abstract
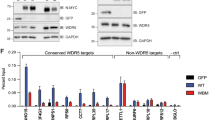
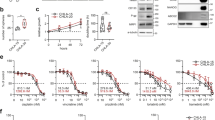
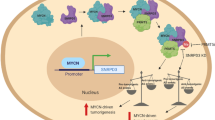
Neuroblastoma is the most common and deadliest tumor in infancy. WDR5 (WD Repeat Domain 5), a critical factor supporting an N-myc transcriptional complex via its WBM site and interacting with chromosome via its WIN site, promotes the progression of neuroblastoma, thus making it a potential anti-neuroblastoma drug target. So far, a few WIN site inhibitors have been reported, and the WBM site disruptors are rare to see. In this study we conducted virtual screening to identify candidate hit compounds targeting the WBM site of WDR5. As a result, 60 compounds were selected as candidate WBM site inhibitors. Cell proliferation assay demonstrated 6 structurally distinct WBM site inhibitors, numbering as compounds 4, 7, 11, 13, 19 and 22, which potently suppressed 3 neuroblastoma cell lines (MYCN-amplified IMR32 and LAN5 cell lines, and MYCN-unamplified SK-N-AS cell line). Among them, compound 19 suppressed the proliferation of IMR32 and LAN5 cells with EC50 values of 12.34 and 14.89 μM, respectively, and exerted a moderate inhibition on SK-N-AS cells, without affecting HEK293T cells at 20 μM. Analysis of high-resolution crystal complex structure of compound 19 against WDR5 revealed that it competitively occupied the hydrophobic pocket where V264 was located, which might disrupt the interaction of MYC with WDR5 and further MYC-medicated gene transcription. By performing RNA-seq analysis we demonstrated the differences in molecular action mechanisms of the compound 19 and a WIN site inhibitor OICR-9429. Most interestingly, we established the particularly high synergy rate by combining WBM site inhibitor 19 and the WIN site inhibitor OICR-9429, providing a novel therapeutic avenue for neuroblastoma.
Similar content being viewed by others
Login or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, et al. Neuroblastoma. Nat Rev Dis Prim. 2016;2:16078.
Twist CJ, Schmidt ML, Naranjo A, London WB, Tenney SC, Marachelian A, et al. Maintaining outstanding outcomes using response- and biology-based therapy for intermediate-risk neuroblastoma: a report from the children’s oncology group study anbl0531. J Clin Oncol. 2019;37:3243–55.
Kholodenko IV, Kalinovsky DV, Doronin, II, Deyev SM, Kholodenko RV. Neuroblastoma origin and therapeutic targets for immunotherapy. J Immunol Res. 2018;2018:7394268. https://doi.org/10.1155/2018/7394268.
Castel V, Cañete A, Navarro S, García-Miguel P, Melero C, Acha T, et al. Outcome of high-risk neuroblastoma using a dose intensity approach: improvement in initial but not in long-term results. Med Pediatr Oncol. 2001;37:537–42.
Migliori V, Mapelli M, Guccione E. On WD40 proteins: propelling our knowledge of transcriptional control? Epigenetics. 2012;7:815–22.
Wang F, Zhang J, Ke X, Peng W, Zhao G, Peng S, et al. WDR5-Myc axis promotes the progression of glioblastoma and neuroblastoma by transcriptional activating CARM1. Biochem Biophys Res Commun. 2020;523:699–706.
Sun Y, Bell JL, Carter D, Gherardi S, Poulos RC, Milazzo G, et al. WDR5 supports an N-myc transcriptional complex that drives a protumorigenic gene expression signature in neuroblastoma. Cancer Res. 2015;75:5143–54.
Thomas LR, Wang Q, Grieb BC, Phan J, Foshage AM, Sun Q, et al. Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC. Mol Cell. 2015;58:440–52.
Thomas LR, Adams CM, Wang J, Weissmiller AM, Creighton J, Lorey SL, et al. Interaction of the oncoprotein transcription factor MYC with its chromatin cofactor WDR5 is essential for tumor maintenance. Proc Natl Acad Sci USA. 2019;116:25260–8.
Rickman DS, Schulte JH, Eilers M. The expanding world of N-myc-driven tumors. Cancer Discov. 2018;8:150–63.
Otte J, Dyberg C, Pepich A, Johnsen JI. MYCN function in neuroblastoma development. Front Oncol. 2021;10:624079.
Ernst P, Vakoc CR. WRAD: enabler of the SET1-family of H3K4 methyltransferases. Brief Funct Genomics. 2012;11:217–26.
Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–70.
Zhao J, Chen W, Pan Y, Zhang Y, Sun H, Wang H, et al. Structural insights into the recognition of histone H3Q5 serotonylation by WDR5. Sci Adv. 2021;7:eabf4291.
Song JJ, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem. 2008;283:35258–64.
Odho Z, Southall SM, Wilson JR. Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. J Biol Chem. 2010;285:32967–76.
Tian J, Teuscher KB, Aho ER, Alvarado JR, Mills JJ, Meyers KM, et al. Discovery and structure-based optimization of potent and selective WD repeat ___domain 5 (WDR5) inhibitors containing a dihydroisoquinolinone bicyclic core. J Med Chem. 2020;63:656–75.
Aho ER, Wang J, Gogliotti RD, Howard GC, Phan J, Acharya P, et al. Displacement of WDR5 from chromatin by a WIN site inhibitor with picomolar affinity. Cell Rep. 2019;26:2916–28.e13.
Wang F, Jeon KO, Salovich JM, Macdonald JD, Alvarado J, Gogliotti RD, et al. Discovery of potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-site inhibitors using fragment-based methods and structure-based design. J Med Chem. 2018;61:5623–42.
Karatas H, Li Y, Liu L, Ji J, Lee S, Chen Y, et al. Discovery of a highly potent, cell-permeable macrocyclic peptidomimetic (MM-589) targeting the WD repeat ___domain 5 protein (WDR5)-mixed lineage leukemia (MLL) protein-protein interaction. J Med Chem. 2017;60:4818–39.
Getlik M, Smil D, Zepeda-Velázquez C, Bolshan Y, Poda G, Wu H, et al. Structure-based optimization of a small molecule antagonist of the interaction between WD repeat-containing protein 5 (WDR5) and mixed-lineage leukemia 1 (MLL1). J Med Chem. 2016;59:2478–96.
Senisterra G, Wu H, Allali-Hassani A, Wasney GA, Barsyte-Lovejoy D, Dombrovski L, et al. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem J. 2013;449:151–9.
Bolshan Y, Getlik M, Kuznetsova E, Wasney GA, Hajian T, Poda G, et al. Synthesis, optimization, and evaluation of novel small molecules as antagonists of WDR5-MLL interaction. CS Med Chem Lett. 2013;4:353–7.
Karatas H, Townsend EC, Cao F, Chen Y, Bernard D, Liu L, et al. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J Am Chem Soc. 2013;135:669–82.
Marschalek R. Mechanisms of leukemogenesis by MLL fusion proteins. Br J Haematol. 2011;152:141–54.
Marschalek R. MLL leukemia and future treatment strategies. Arch Pharm (Weinheim). 2015;348:221–8.
Chen IJ, Foloppe N. Drug-like bioactive structures and conformational coverage with the LigPrep/ConfGen suite: comparison to programs MOE and catalyst. J Chem Inf Model. 2010;50:822–39.
Chacón Simon S, Wang F, Thomas LR, Phan J, Zhao B, Olejniczak ET, et al. Discovery of WD repeat-containing protein 5 (WDR5)-MYC inhibitors using fragment-based methods and structure-based design. J Med Chem. 2020;63:4315–33.
Macdonald JD, Chacón Simon S, Han C, Wang F, Shaw JG, Howes JE, et al. Discovery and optimization of salicylic acid-derived sulfonamide inhibitors of the WD repeat-containing protein 5-MYC protein-protein interaction. J Med Chem. 2019;62:11232–59.
Liu X, Jiang H, Li H. SHAFTS: A hybrid approach for 3D molecular similarity calculation. 1. Method and assessment of virtual screening. J Chem Inf Model. 2011;51:2372–85.
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49.
Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–66.
Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–42.
Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32.
Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54.
Ianevski A, Giri AK, Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020;48:W488–W93.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323.
Costa-Silva J, Domingues D, Lopes FM. RNA-Seq differential expression analysis: An extended review and a software tool. PLoS One. 2017;12:e0190152.
da Silva Rocha SFL, Olanda CG, Fokoue HH, Sant’Anna CMR. Virtual screening techniques in drug discovery: review and recent applications. Curr Top Med Chem. 2019;19:1751–67.
Ferreira LG, Dos Santos RN, Oliva G, Andricopulo AD. Molecular docking and structure-based drug design strategies. Molecules. 2015;20:13384–421.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Wang X, Simpson ER, Brown KA. p53: Protection against tumor growth beyond effects on cell cycle and apoptosis. Cancer Res. 2015;75:5001–7.
Bliss CI. The calculation of microbial assays. Bacteriol Rev. 1956;20:243–58.
Guarnaccia AD, Tansey WP. Moonlighting with WDR5: A cellular multitasker. J Clin Med. 2018;7:21.
Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell. 2017;170:564–76.
Wu W, Yu N, Li F, Gao P, Lin S, Zhu Y. RPL35 promotes neuroblastoma progression via the enhanced aerobic glycolysis. Am J Cancer Res. 2021;11:5701–14.
Zheng J, Lang Y, Zhang Q, Cui D, Sun H, Jiang L, et al. Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation. Genes Dev. 2015;29:1524–34.
Lu F, Wu X, Yin F, Chia-Fang Lee C, Yu M, Mihaylov IS, et al. Regulation of DNA replication and chromosomal polyploidy by the MLL-WDR5-RBBP5 methyltransferases. Biol Open. 2016;5:1449–60.
Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1:1001–8.
Acknowledgements
This study was sponsored by Shanghai Municipal Science and Technology Commission (21Y11912200 to Kai Li), the Cyrus Tang Foundation, Clinical Research Plan of SHDC (SHDC2020CR2009A), Shanghai Municipal Key Clinical Specialty (shslczdzk05703), Hengjie Special Support Plan (2022 to Kai Li), Lingang Laboratory (LG202102-01-03), National Natural Science Foundation of China (82003654), Shanghai Science and Technology Development Funds (20QA1406400), and the Science and Technology Commission of Shanghai Municipality Grants (20431900100, 2043190012 and 20430780300); as well as the startup package from ShanghaiTech University. We also thank the HPC Platform of ShanghaiTech University and Shanghai Supercomputer Center for computing time.
Author information
Authors and Affiliations
Contributions
FB, KL, and LHM conceptualized and designed the study. PXR and LW performed virtual screening. QLH, XLZ, YC, and WHL performed molecular biology experiments and analyzed the data. QLH and XLZ drafted the manuscript, which was revised and embellished by FB. FB, and KL supervised the study. All authors read and approved the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, Ql., Zhang, Xl., Ren, Px. et al. Discovery, evaluation and mechanism study of WDR5-targeted small molecular inhibitors for neuroblastoma. Acta Pharmacol Sin 44, 877–887 (2023). https://doi.org/10.1038/s41401-022-00999-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-022-00999-z
Keywords
This article is cited by
-
MAI-TargetFisher: A proteome-wide drug target prediction method synergetically enhanced by artificial intelligence and physical modeling
Acta Pharmacologica Sinica (2025)
-
The NTE ___domain of PTENα/β promotes cancer progression by interacting with WDR5 via its SSSRRSS motif
Cell Death & Disease (2024)
-
DeepSA: a deep-learning driven predictor of compound synthesis accessibility
Journal of Cheminformatics (2023)
-
WDR5 facilitates recruitment of N-MYC to conserved WDR5 gene targets in neuroblastoma cell lines
Oncogenesis (2023)