Abstract
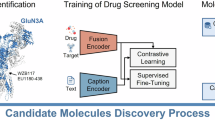
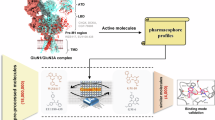
The GluN1/GluN3A receptor, a unique excitatory glycine receptor recently identified in the central nervous system, challenges traditional perspectives of N-methyl-D-aspartate (NMDA) receptor diversity and glycinergic signaling. Its role in emotional regulation positions it as a potential therapeutic target for neuropsychiatric disorders. However, pharmacological research on GluN1/GluN3A receptors remains at an early stage. Traditional high-throughput screening methods for ion channel drug discovery often lack efficiency, particularly when applied to large compound libraries. To address this concern, we designed a deep learning-based strategy that balances efficiency and accuracy for identifying GluN1/GluN3A inhibitors. First, a sequence-based scoring function was developed to rapidly screen a library containing 18 million compounds, reducing the pool to approximately 105 candidates. Next, two complex-based scoring functions, IGModel and RTMScore, were employed to precisely score and rank the remaining candidates. Finally, an active molecule with an IC50 of 2.87 ± 0.80 μM for the GluN1/GluN3A receptor was confirmed through whole-cell voltage-clamp electrophysiology. This study also presents a paradigm for integrating deep learning into rapid and precise high-throughput screening.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hansen KB, Yi F, Perszyk RE, Furukawa H, Wollmuth LP, Gibb AJ, et al. Structure, function, and allosteric modulation of NMDA receptors. J Gen Physiol. 2018;150:1081–105.
Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–96.
Newcomer JW, Farber NB, Olney JW. NMDA receptor function, memory, and brain aging. Dialogues Clin Neurosci. 2000;2:219–32.
Grand T, Abi Gerges S, David M, Diana MA, Paoletti P. Unmasking GluN1/GluN3A excitatory glycine NMDA receptors. Nat Commun. 2018;9:4769.
Galliano E, Schonewille M, Peter S, Rutteman M, Houtman S, Jaarsma D, et al. Impact of NMDA receptor overexpression on cerebellar Purkinje cell activity and motor learning. eNeuro. 2018;5:ENEURO.0270-17.2018.
Karakas E, Furukawa H. Crystal structure ofa heterotetrameric NMDA receptor ion channel. Science. 2014;344:992–7.
Ma T, Cheng Q, Chen C, Luo Z, Feng D. Excessive activation of NMDA receptors in the pathogenesis of multiple peripheral organs via mitochondrial dysfunction, oxidative stress, and inflammation. SN Compr Clin Med. 2020;2:551–69.
Parsons CG, Danysz W, Quack G. Memantine is a clinically well tolerated N-methyl-D-aspartate (NMDA) receptor antagonist—a review of preclinical data. Neuropharmacology. 1999;38:735–67.
Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, et al. Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature. 2002;415:793–8.
Bossi S, Pizzamiglio L, Paoletti P. Excitatory GluN1/GluN3A glycine receptors (eGlyRs) in brain signaling. Trends Neurosci. 2023;46:667–81.
Zeng Y, Zheng Y, Zhang T, Ye F, Zhan L, Kou Z, et al. Identification of a subtype-selective allosteric inhibitor of GluN1/GluN3 NMDA receptors. Front Pharmacol. 2022; 13.
Wang W, Gao X. Deep learning in bioinformatics. Methods 2019;166:1–3.
Vamathevan J, Clark D, Czodrowski P, Dunham I, Ferran E, Lee G, et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18:463–77.
Lavecchia A. Deep learning in drug discovery: opportunities, challenges and future prospects. Drug Discov Today. 2019;24:2017–32.
Lo YC, Rensi SE, Torng W, Altman RB. Machine learning in chemoinformatics and drug discovery. Drug Discov Today. 2018;23:1538–46.
Carpenter KA, Cohen DS, Jarrell JT, Huang X. Deep learning and virtual drug screening. Future Med Chem. 2018;10:2557–67.
Oliveira T, Silva M, Maia E, Silva A, Taranto A. Virtual screening algorithms in drug discovery: a review focused on machine and deep learning methods. Drugs Drug Candidates. 2023;2:311–34.
Koh HY, Nguyen ATN, Pan S, May LT, Webb GI. Physicochemical graph neural network for learning protein–ligand interaction fingerprints from sequence data. Nat Mach Intell. 2024;6:673–87.
Ozturk H, Ozgur A, Ozkirimli E. DeepDTA: deep drug-target binding affinity prediction. Bioinformatics. 2018;34:i821–i9.
Nguyen T, Le H, Quinn TP, Nguyen T, Le TD, Venkatesh S. GraphDTA: predicting drug-target binding affinity with graph neural networks. Bioinformatics. 2021;37:1140–7.
Bai P, Miljković F, John B, Lu H. Interpretable bilinear attention network with ___domain adaptation improves drug–target prediction. Nat Mach Intell. 2023;5:126–36.
Lu W, Wu Q, Zhang J, Rao J, Li C, Zheng S. Tankbind: Trigonometry-aware neural networks for drug-protein binding structure prediction. Adv Neural Inf Process Syst. 2022;35:7236–49.
Zheng L, Fan J, Mu Y. OnionNet: a multiple-layer intermolecular-contact-based convolutional neural network for protein-ligand binding affinity prediction. ACS Omega. 2019;4:15956–65.
Wang Z, Zheng L, Liu Y, Qu Y, Li YQ, Zhao M, et al. OnionNet-2: A convolutional neural network model for predicting protein-ligand binding affinity based on residue-atom contacting shells. Front Chem. 2021;9:753002.
Nguyen DD, Wei GW. AGL-score: algebraic graph learning score for protein-ligand binding scoring, ranking, docking, and screening. J Chem Inf Model. 2019;59:3291–304.
Wang Z, Wang S, Li Y, Guo J, Wei Y, Mu Y, et al. A new paradigm for applying deep learning to protein-ligand interaction prediction. Brief Bioinform. 2024;25:bbae145.
Wang Z, Zheng L, Wang S, Lin M, Wang Z, Kong AW, et al. A fully differentiable ligand pose optimization framework guided by deep learning and a traditional scoring function. Brief Bioinform. 2023;24:bbac520.
Zheng L, Meng J, Jiang K, Lan H, Wang Z, Lin M, et al. Improving protein-ligand docking and screening accuracies by incorporating a scoring function correction term. Brief Bioinform. 2022;23:bbac051.
Shen T, Liu F, Wang Z, Sun J, Bu Y, Meng J, et al. zPoseScore model for accurate and robust protein-ligand docking pose scoring in CASP15. Proteins. 2023;91:1837–49.
Méndez-Lucio O, Ahmad M, del Rio-Chanona EA, Wegner JK. A geometric deep learning approach to predict binding conformations of bioactive molecules. Nat Mach Intell. 2021;3:1033–9.
Shen C, Zhang X, Deng Y, Gao J, Wang D, Xu L, et al. Boosting protein-ligand binding pose prediction and virtual screening based on residue-atom distance likelihood potential and graph transformer. J Med Chem. 2022;65:10691–706.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9.
Lin Z, Akin H, Rao R, Hie B, Zhu Z, Lu W, et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science. 2023;379:1123–30.
Edwards C, Lai T, Ros K, Honke G, Cho K, Ji H. Translation between molecules and natural language. Preprint at https://arxiv.org/abs/2204.11817.
Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–201.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinforma. 2009;10:421.
Yan X, Lu Y, Li Z, Wei Q, Gao X, Wang S, et al. PointSite: A point cloud segmentation tool for identification of protein ligand binding atoms. J Chem Inf Model. 2022;62:2835–45.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61.
Elnaggar A, Heinzinger M, Dallago C, Rehawi G, Wang Y, Jones L, et al. ProtTrans: Toward understanding the language of life through self-supervised learning. IEEE Trans Pattern Anal Mach Intell. 2022;44:7112–27.
Pei Q, Wu L, Zhu J, Xia Y, Xie S, Qin T, et al. Breaking the barriers of data scarcity in drug-target affinity prediction. Brief Bioinform. 2023;24:bbad386.
Meda RS, Farimani AB. BAPULM: Binding affinity prediction using language models. Preprint at https://arxiv.org/abs/2411.04150.
Su M, Yang Q, Du Y, Feng G, Liu Z, Li Y, et al. Comparative assessment of scoring functions: The CASF-2016 Update. J Chem Inf Model. 2019;59:895–913.
Zhu Z, Yi F, Epplin MP, Liu D, Summer SL, Mizu R, et al. Negative allosteric modulation of GluN1/GluN3 NMDA receptors. Neuropharmacology. 2020;176:108117.
Crawley O, Conde-Dusman MJ, Perez-Otano I. GluN3A NMDA receptor subunits: more enigmatic than ever? J Physiol. 2022;600:261–76.
Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold3. Nature. 2024;630:493–500.
Acknowledgements
This research is supported by the Taishan Scholars Program of Shandong Province (tstp20240807), Shandong Provincial Natural Science Foundation (ZR2024MA071) and the National Science and Technology Innovation 2030 Major Program (2021ZD0200900). The authors sincerely thank the support from the Core Facility Sharing Platform of Shandong University and the National Demonstration Center for Experimental Physics Education (Shandong University).
Author information
Authors and Affiliations
Contributions
WFL and ZBG designed the study. ZCW and JYS designed the deep learning-based screening strategy and conducted high-throughput virtual screening. YZ, XQC, and HCW designed and conducted wet-lab experiments. YYL, YGM, and LZZ analyzed the data. ZCW and YZ wrote the manuscript. All authors reviewed the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Zc., Zeng, Y., Sun, Jy. et al. An efficient deep learning-based strategy to screen inhibitors for GluN1/GluN3A receptor. Acta Pharmacol Sin (2025). https://doi.org/10.1038/s41401-025-01513-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41401-025-01513-x