Abstract
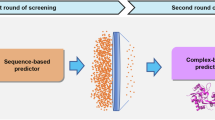
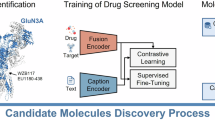
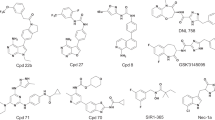
Ligand-based drug discovery methods typically utilize pharmacophore similarities among molecules to screen for potential active compounds. Among these, scaffold hopping is a widely used ligand-based lead identification strategy that facilitates clinical candidate discovery by seeking inhibitors with similar biological activity yet distinct scaffolds. In this study, we employed GeminiMol, a deep learning-based molecular representation framework that incorporates bioactive conformational space information. This approach enables ligand-based virtual screening by referencing known active compounds to identify potential hits with similar structural and bioactive conformational features. Using GeminiMol-based ligand screening method, we discovered a potent GluN1/GluN3A inhibitor, GM-10, from an 18-million-compound library. Notably, GM-10 features a completely different scaffold compared to known inhibitors. Subsequent validation using whole-cell patch-clamp recording confirmed its activity, with an IC50 of 0.98 ± 0.13 μM for GluN1/GluN3A. Further optimization is required to enhance its selectivity, as it exhibited IC50 values of 3.89 ± 0.79 μM for GluN1/GluN2A and 1.03 ± 0.21 μM for GluN1/GluN3B. This work highlights the potential of AI-driven molecular representation technologies to facilitate scaffold hopping and enhance similarity-based virtual screening for drug discovery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hansen KB, Wollmuth LP, Bowie D, Furukawa H, Menniti FS, Sobolevsky AI, et al. Structure, function, and pharmacology of glutamate receptor ion channels. Pharmacol Rev. 2021;73:298–487.
Hansen KB, Yi F, Perszyk RE, Furukawa H, Wollmuth LP, Gibb AJ, et al. Structure, function, and allosteric modulation of NMDA receptors. J Gen Physiol 2018;150:1081–105.
Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383–400.
Pérez-Otaño I, Schulteis CT, Contractor A, Lipton SA, Trimmer JS, Sucher NJ, et al. Assembly with the NR1 subunit is required for surface expression of NR3A-containing NMDA receptors. J Neurosci. 2001;21:1228–37.
Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, et al. Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature. 2002;415:793–8.
Bendel O, Meijer B, Hurd Y, von Euler G. Cloning and expression of the human NMDA receptor subunit NR3B in the adult human hippocampus. Neurosci Lett 2005;377:31–6.
Sucher NJ, Akbarian S, Chi CL, Leclerc CL, Awobuluyi M, Deitcher DL, et al. Developmental and regional expression pattern of a novel NMDA receptor-like subunit (NMDAR-L) in the rodent brain. J Neurosci 1995;15:6509–20.
Perez-Otano I, Larsen RS, Wesseling JF. Emerging roles of GluN3-containing NMDA receptors in the CNS. Nat Rev Neurosci. 2016;17:623–35.
Das S, Sasaki YF, Rothe T, Premkumar LS, Takasu M, Crandall JE, et al. Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature. 1998;393:377–81.
Bossi S, Pizzamiglio L, Paoletti P. Excitatory GluN1/GluN3A glycine receptors (eGlyRs) in brain signaling. Trends Neurosci. 2023;46:667–81.
Marco S, Giralt A, Petrovic MM, Pouladi MA, Martinez-Turrillas R, Martinez-Hernandez J, et al. Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington’s disease models. Nat Med. 2013;19:1030–8.
Beesley S, Sullenberger T, Crotty K, Ailani R, D’Orio C, Evans K, et al. D-serine mitigates cell loss associated with temporal lobe epilepsy. Nat Commun. 2020;11:4966.
Mueller HT, Meador-Woodruff JH. NR3A NMDA receptor subunit mRNA expression in schizophrenia, depression and bipolar disorder. Schizophr Res. 2004;71:361–70.
Otsu Y, Darcq E, Pietrajtis K, Matyas F, Schwartz E, Bessaih T, et al. Control of aversion by glycine-gated GluN1/GluN3A NMDA receptors in the adult medial habenula. Science. 2019;366:250–4.
Yuan T, Mameli M, O’Connor EC, Dey PN, Verpelli C, Sala C, et al. Expression of cocaine-evoked synaptic plasticity by GluN3A-containing NMDA receptors. Neuron. 2013;80:1025–38.
Zhong W, Wu A, Berglund K, Gu X, Jiang MQ, Talati J, et al. Pathogenesis of sporadic Alzheimer’s disease by deficiency of NMDA receptor subunit GluN3A. Alzheimers Dement. 2022;18:222–39.
Yao Y, Mayer ML. Characterization of a soluble ligand binding ___domain of the NMDA receptor regulatory subunit NR3A. J Neurosci. 2006;26:4559–66.
Grand T, Abi Gerges S, David M, Diana MA, Paoletti P. Unmasking GluN1/GluN3A excitatory glycine NMDA receptors. Nat Commun. 2018;9:4769.
Kvist T, Greenwood JR, Hansen KB, Traynelis SF, Brauner-Osborne H. Structure-based discovery of antagonists for GluN3-containing N-methyl-D-aspartate receptors. Neuropharmacology. 2013;75:324–36.
Zeng Y, Zheng Y, Zhang T, Ye F, Zhan L, Kou Z, et al. Identification of a subtype-selective allosteric inhibitor of GluN1/GluN3 NMDA receptors. Front Pharmacol. 2022;13:888308.
Zhu Z, Yi F, Epplin MP, Liu D, Summer SL, Mizu R, et al. Negative allosteric modulation of GluN1/GluN3 NMDA receptors. Neuropharmacology. 2020;176:108117.
Cummings KA, Popescu GK. Protons potentiate GluN1/GluN3A currents by attenuating their desensitisation. Sci Rep. 2016;6:23344.
Hemelikova K, Kolcheva M, Skrenkova K, Kaniakova M, Horak M. Lectins modulate the functional properties of GluN1/GluN3-containing NMDA receptors. Neuropharmacology. 2019;157:107671.
Geppert H, Vogt M, Bajorath J. Current trends in ligand-based virtual screening: molecular representations, data mining methods, new application areas, and performance evaluation. J Chem Inf Model. 2010;50:205–16.
Ripphausen P, Nisius B, Bajorath J. State-of-the-art in ligand-based virtual screening. Drug Discov Today. 2011;16:372–6.
da Rocha MN, de Sousa DS, da Silva Mendes FR, dos Santos HS, Marinho GS, Marinho MM, et al. Ligand and structure-based virtual screening approaches in drug discovery: minireview. Mol Divers. 2024. https://doi.org/10.1007/s11030-024-10979-6.
Willett P. The calculation of molecular structural similarity: principles and practice. Mol Inf. 2014;33:403–13.
Zhang Q, Muegge I. Scaffold hopping through virtual screening using 2D and 3D similarity descriptors: Ranking, voting, and consensus scoring. J Med Chem. 2006;49:1536–48.
Wang L, Wang S, Yang H, Li S, Wang X, Zhou Y, et al. Conformational space profiling enhances generic molecular representation for AI-powered ligand-based drug discovery. Adv Sci. 2024;11:e2403998.
Wang S, Han Q, Qin W, Wang L, Yuan J, Zhao Y, et al. PhenoScreen: a dual-space contrastive learning framework-based phenotypic screening method by linking chemical perturbations to cellular morphology. bioRxiv. 2024.
Wu H, Liu J, Zhang R, Lu Y, Cui G, Cui Z, et al. A review of deep learning methods for ligand based drug virtual screening. Fundam Res. 2024;4:715–37.
Ogden KK, Traynelis SF. Contribution of the M1 transmembrane helix and pre-M1 region to positive allosteric modulation and gating of N-methyl-D-aspartate receptors. Mol Pharmacol. 2013;83:1045–56.
Chou TH, Epstein M, Michalski K, Fine E, Biggin PC, Furukawa H. Structural insights into binding of therapeutic channel blockers in NMDA receptors. Nat Struct Mol Biol. 2022;29:507–18.
Rouzbeh N, Rau AR, Benton AJ, Yi F, Anderson CM, Johns MR, et al. Allosteric modulation of GluN1/GluN3 NMDA receptors by GluN1-selective competitive antagonists. J Gen Physiol 2023;155:e202313340.
Wang TM, Brown BM, Deng L, Sellers BD, Lupardus PJ, Wallweber HJA, et al. A novel NMDA receptor positive allosteric modulator that acts via the transmembrane ___domain. Neuropharmacology. 2017;121:204–18.
Talukder I, Borker P, Wollmuth LP. Specific sites within the ligand-binding ___domain and ion channel linkers modulate NMDA receptor gating. J Neurosci. 2010;30:11792–804.
Hofner G, Hoesl CE, Parsons C, Quack G, Wanner KT. NMDA-NR2B subtype selectivity of stereoisomeric 2-(1,2,3,4-tetrahydro-1-isoquinolyl)ethanol derivatives. Bioorg Med Chem Lett. 2005;15:2231–4.
Zhang Y, Ye F, Zhang T, Lv S, Zhou L, Du D, et al. Structural basis of ketamine action on human NMDA receptors. Nature. 2021;596:301–5.
Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–96.
Chou TH, Tajima N, Romero-Hernandez A, Furukawa H. Structural basis of functional transitions in mammalian NMDA receptors. Cell. 2020;182:357–71.e313.
Sastry GM, Dixon SL, Sherman W. Rapid shape-based ligand alignment and virtual screening method based on atom/feature-pair similarities and volume overlap scoring. J Chem Inf Model. 2011;51:2455–66.
Acknowledgements
This work was supported by Shanghai Science and Technology Development Funds (Grant IDs: 24JS2850100 and 24JS2850200), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant ID: XDB0830403), ShanghaiTech AI4S Initiative SHTAI4S202404, National Key R&D Program of China (Grant IDs: 2022YFC3400501 & 2022YFC3400500), start-up package from ShanghaiTech University, and Shanghai Frontiers Science Center for Biomacromolecules and Precision Medicine at ShanghaiTech University, and the National Science and Technology Innovation 2030 Major Program (Grant ID: 2021ZD0200900). The authors appreciate the technical support provided by the high-performance computing cluster of ShanghaiTech University.
Author information
Authors and Affiliations
Contributions
FB and ZBG contributed to conceptualization. SHW and LW contributed to model development. SHW, HY, YQZ and SYT contributed to data curation. YZ, XQC and HYW designed and conducted wet-lab experiments. SHW, YZ, HY, YQZ, SYT, LW, ZBG, and FB contributed to original draft writing. All authors reviewed the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Sh., Zeng, Y., Yang, H. et al. Discovery of novel GluN1/GluN3A NMDA receptor inhibitors using a deep learning-based method. Acta Pharmacol Sin (2025). https://doi.org/10.1038/s41401-025-01571-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41401-025-01571-1