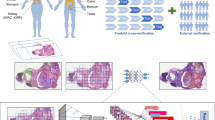
Abstract
Background
The aim of this study was to construct a clinically practical model to precisely predict lymph node (LN) metastasis in bladder cancer patients.
Methods
Four independent cohorts were included. The least absolute shrinkage and selection operator regression with multivariate logistic regression were applied. The diagnostic efficacy of LN score and CT/MRI was compared by accuracy, sensitivity, specificity, and area under curve (AUC).
Results
A total of 606 patients were included to develop a basic prediction model. After multistep gene selection, the LN metastasis prediction model was constructed with 5 genes. The model can accurately predict LN metastasis with an AUC of 0.781. For clinically practical use, we transformed the model into a Fast LN Scoring System using the SYSMH cohort (n = 105). High LN score patients exhibited a 72.2% LN metastasis rate, while low LN score patients showed a 3.4% LN metastasis rate. The LN score achieved a superior accuracy than CT/MRI (0.882 vs. 0.727). Application of LN score can correct the diagnosis of 88% (22/25) patients who were misdiagnosed by CT/MRI.
Discussion
The clinically practical LN score can precisely, rapidly, and conveniently predict LN status, which will assist preoperative diagnosis for LN metastasis and guide precise therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
269,00 € per year
only 11,21 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The raw RNA-seq data can be acquired from the TCGA database (The Cancer Genome Atlas, https://xenabrowser.net/datapages/) and GEO database (Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/). The anonymous patient characteristics and qPCR results can be obtained in the Supplementary Materials.
Code availability
The code used in this study can be obtained at https://github.com/JunlinLu123/LNscore.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Chen X, Zhang J, Ruan W, Huang M, Wang C, Wang H, et al. Urine DNA methylation assay enables early detection and recurrence monitoring for bladder cancer. J Clin Investig. 2020;130:6278–89.
Liu S, Chen X, Lin T. Emerging strategies for the improvement of chemotherapy in bladder cancer: current knowledge and future perspectives. J Adv Res. 2022;39:187–202.
Zhang Q, Liu S, Wang H, Xiao K, Lu J, Chen S, et al. ETV4 mediated tumor-associated neutrophil infiltration facilitates lymphangiogenesis and lymphatic metastasis of bladder cancer. Adv Sci. 2023;10:e2205613.
Youssef RF, Raj GV. Lymphadenectomy in management of invasive bladder cancer. Int J Surg Oncol. 2011;2011:758189.
Karl A, Carroll PR, Gschwend JE, Knüchel R, Montorsi F, Stief CG, et al. The impact of lymphadenectomy and lymph node metastasis on the outcomes of radical cystectomy for bladder cancer. Eur Urol. 2009;55:826–35.
Del Bene G, Calabrò F, Giannarelli D, Plimack ER, Harshman LC, Yu EY, et al. Neoadjuvant vs. adjuvant chemotherapy in muscle invasive bladder cancer (MIBC): analysis from the RISC database. Front Oncol. 2018;8:463.
McKibben MJ, Woods ME. Preoperative imaging for staging bladder cancer. Curr Urol Rep. 2015;16:22.
Liu S, Chen X, Lin T. Lymphatic metastasis of bladder cancer: molecular mechanisms, diagnosis and targeted therapy. Cancer Lett. 2021;505:13–23.
Huang M, Dong W, Xie R, Wu J, Su Q, Li W, et al. HSF1 facilitates the multistep process of lymphatic metastasis in bladder cancer via a novel PRMT5-WDR5-dependent transcriptional program. Cancer Commun. 2022;42:447–70.
Xie R, Chen X, Cheng L, Huang M, Zhou Q, Zhang J, et al. NONO inhibits lymphatic metastasis of bladder cancer via alternative splicing of SETMAR. Mol Ther. 2021;29:291–307.
Luo C, Huang B, Wu Y, Xu Y, Ou W, Chen J, et al. Identification of lymph node metastasis-related key genes and prognostic risk model in bladder cancer by co-expression analysis. Front Mol Biosci. 2021;8:633299.
Cao R, Ma B, Wang G, Xiong Y, Tian Y, Yuan L. An epithelial-mesenchymal transition (EMT) preoperative nomogram for prediction of lymph node metastasis in bladder cancer (BLCA). Dis Markers. 2020;2020:8833972.
Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–5.
Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–95.
Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77.
Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Mak. 2006;26:565–74.
Peng S, Chen X, Huang C, Yang C, Situ M, Zhou Q, et al. UBE2S as a novel ubiquitinated regulator of p16 and β-catenin to promote bone metastasis of prostate cancer. Int J Biol Sci. 2022;18:3528–43.
Chen X, Xie R, Gu P, Huang M, Han J, Dong W, et al. Long noncoding RNA LBCS inhibits self-renewal and chemoresistance of bladder cancer stem cells through epigenetic silencing of SOX2. Clin Cancer Res. 2019;25:1389–403.
Wang B, Wan F, Sheng H, Zhu Y, Shi G, Zhang H, et al. Identification and validation of an 18-gene signature highly-predictive of bladder cancer metastasis. Sci Rep. 2018;8:374.
Smith SC, Baras AS, Dancik G, Ru Y, Ding KF, Moskaluk CA, et al. A 20-gene model for molecular nodal staging of bladder cancer: development and prospective assessment. Lancet Oncol. 2011;12:137–43.
Liu J, Zheng Z, Zhang W, Wan M, Ma W, Wang R, et al. Dysregulation of tumor microenvironment promotes malignant progression and predicts risk of metastasis in bladder cancer. Ann Transl Med. 2021;9:1438.
Lu X, Wang Y, Jiang L, Gao J, Zhu Y, Hu W, et al. A pre-operative nomogram for prediction of lymph node metastasis in bladder urothelial carcinoma. Front Oncol. 2019;9:488.
Dobrocky T, Grunder L, Breiding PS, Branca M, Limacher A, Mosimann PJ, et al. Assessing spinal cerebrospinal fluid leaks in spontaneous intracranial hypotension with a scoring system based on brain magnetic resonance imaging findings. JAMA Neurol. 2019;76:580–7.
Lin KC, Chen TM, Yuan KS, Wu ATH, Wu SY. Assessment of predictive scoring system for 90-day mortality among patients with locally advanced head and neck squamous cell carcinoma who have completed concurrent chemoradiotherapy. JAMA Netw Open. 2020;3:e1920671.
Wang L, Yang C, Li F, Mu D, Ran P, Shen H, et al. High levels of MESP1 expression in non-small cell lung cancer can facilitate cell proliferation, metastasis and suppresses cell apoptosis. Transl Cancer Res. 2020;9:5956–68.
Yin X, Fang S, Wang M, Wang Q, Fang R, Chen J. EFEMP1 promotes ovarian cancer cell growth, invasion and metastasis via activated the AKT pathway. Oncotarget. 2016;7:47938–53.
Zhang D, Han S, Pan X, Li H, Zhao H, Gao X, et al. EFEMP1 binds to STEAP1 to promote osteosarcoma proliferation and invasion via the Wnt/β-catenin and TGF-β/Smad2/3 signal pathways. J Bone Oncol. 2022;37:100458.
Gao X, Yang J. Identification of genes related to clinicopathological characteristics and prognosis of patients with colorectal cancer. DNA Cell Biol. 2020;39:690–9.
Yang B, Li M, Tang W, Liu W, Zhang S, Chen L, et al. Dynamic network biomarker indicates pulmonary metastasis at the tipping point of hepatocellular carcinoma. Nat Commun. 2018;9:678.
Zhou Y, Fu X, Zheng Z, Ren Y, Zheng Z, Zhang B, et al. Identification of gene co-expression modules and hub genes associated with the invasiveness of pituitary adenoma. Endocrine. 2020;68:377–89.
Wu YM, Sa Y, Guo Y, Li QF, Zhang N. Identification of WHO II/III gliomas by 16 prognostic-related gene signatures using machine learning methods. Curr Med Chem. 2022;29:1622–39.
Funding
This study was supported by the National Key Research and Development Program of China (Grant No. 2022YFC2408300), the National Natural Science Foundation of China (Grant Nos. 82273421, 81825016, 82072827, U21A20383), Guangdong Basic and Applied Basic Research Foundation (Grant No. 2021B1515020009), Key Research and Development Program of Guangdong (Grant No. 2018B010109006), Science and Technology Program of Guangzhou (Grant No. 2023A03J0718, 202102010002), Fundamental Research Funds for the Central Universities, Sun Yat-sen University (23ykbj002 for XC), Guangdong Provincial Clinical Research Center for Urological Diseases (2020B1111170006), and Guangdong Science and Technology Department (2020B1212060018, 2018B030317001, 2017B030314026).
Author information
Authors and Affiliations
Contributions
J Lu and XC conceived and designed the study. J Lai and KX downloaded and maintained the public data. J Lu and QX performed bioinformatics analysis. SP and YZ collected patient samples. J Lai conducted PCR. J Lu, XC, and TL wrote the first draft. YC, QZ, and SL reviewed and revised the manuscript. XC and TL provided fundings for the research. All authors read and approved the final version of the manuscript. The authorship order among the co-first authors was assigned based on their relative contributions.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The ethical consent of this study was approved by Sun Yat‐sen University Committees for Ethical Review of Research involving Human Subjects. All human tissue samples were obtained from patients with written informed consent.
Consent for publication
The consent for publication has been acquired from the participants.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, J., Lai, J., Xiao, K. et al. A clinically practical model for the preoperative prediction of lymph node metastasis in bladder cancer: a multicohort study. Br J Cancer 129, 1166–1175 (2023). https://doi.org/10.1038/s41416-023-02383-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02383-y