Abstract
Background
Gastric cancer (GC) is a leading cause of cancer-related deaths worldwide, with late-stage diagnoses frequently leading to poor outcomes. This underscores the need for effective early-stage gastric cancer (ESGC) diagnostics.
Methods
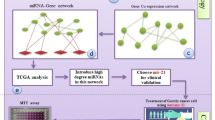
We introduce ESGCmiRD, an innovative artificial intelligence-driven strategy that identifies a miRNA signature for ESGC detection by integrating robust expression patterns, ESGC relevance, and regulatory capabilities of microRNA (miRNA) based on multiple networks. Expression and biological roles of miRNAs in GC were validated and explored via bioinformatics analysis and in vitro studies. miRNA-target interaction was confirmed by dual-luciferase reporter assay. Molecular docking predicted miRNA-drug binding affinities, assessing the miRNA signature’s therapeutic potential.
Results
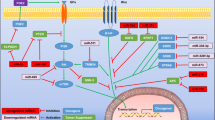
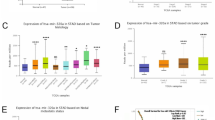
ESGCmiRD identified a blood miRNA signature (miR-320b, miR-222-3p, miR-181a-5p, miR-103a-3p, miR-107) for ESGC detection, demonstrated high diagnostic accuracy with AUC values of 0.986, 0.977, 0.815, and 0.811 in the test and three validation sets (GSE211692, TCGA-STAD, and our cohort), respectively. The five miRNAs were overexpressed in ESGC plasma and directly target PTEN, promoting GC cell proliferation, migration, and invasion. Molecular docking suggested Paclitaxel had the strongest potential interaction with these miRNAs.
Conclusion
This method identifies a robust miRNA signature for ESGC detection and sheds light on gastric carcinogenesis mechanisms, opening doors for potential therapeutic strategies.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
269,00 € per year
only 11,21 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
Data availability
All data generated or analysed are included in this article and its supplementary information files. The code can be found at https://github.com/ljc7878/ESGCmiRD.git.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Li Y, Feng A, Zheng S, Chen C, Lyu J. Recent estimates and predictions of 5-year survival in patients with gastric cancer: a model-based period analysis. Cancer Control: J Moffitt Cancer Cent. 2022;29:10732748221099227.
Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–48.
Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. Cancer J Clin. 2021;71:264–79.
Huang Z-B, Zhang H-T, Yu B, Yu D-H. Cell-free DNA as a liquid biopsy for early detection of gastric cancer. Oncol Lett. 2021;21:3.
Ma S, Zhou M, Xu Y, Gu X, Zou M, Abudushalamu G, et al. Clinical application and detection techniques of liquid biopsy in gastric cancer. Mol Cancer. 2023;22:7.
Guo X, Peng Y, Song Q, Wei J, Wang X, Ru Y, et al. A liquid biopsy signature for the early detection of gastric cancer in patients. Gastroenterology. 2023;165:402–13.e13.
Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic—implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18:297–312.
Lone SN, Nisar S, Masoodi T, Singh M, Rizwan A, Hashem S, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022;21:79.
Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol. 2022;15:131.
Alix-Panabières C, Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discov. 2021;11:858–73.
Zhang Z, Wu H, Chong W, Shang L, Jing C, Li L. Liquid biopsy in gastric cancer: predictive and prognostic biomarkers. Cell Death Dis. 2022;13:903.
Farasati Far B, Vakili K, Fathi M, Yaghoobpoor S, Bhia M, Naimi-Jamal MR. The role of microRNA-21 (miR-21) in pathogenesis, diagnosis, and prognosis of gastrointestinal cancers: a review. Life Sci. 2023;316:121340.
Guo T, Tang X-H, Gao X-Y, Zhou Y, Jin B, Deng Z-Q, et al. A liquid biopsy signature of circulating exosome-derived mRNAs, miRNAs and lncRNAs predict therapeutic efficacy to neoadjuvant chemotherapy in patients with advanced gastric cancer. Mol Cancer. 2022;21:216.
Yan W, Chen Y, Hu G, Shi T, Liu X, Li J, et al. MiR-200/183 family-mediated module biomarker for gastric cancer progression: an AI-assisted bioinformatics method with experimental functional survey. J Transl Med. 2023;21:163.
So JBY, Kapoor R, Zhu F, Koh C, Zhou L, Zou R, et al. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut. 2021;70:829–37.
Abe S, Matsuzaki J, Sudo K, Oda I, Katai H, Kato K, et al. A novel combination of serum microRNAs for the detection of early gastric cancer. Gastric Cancer. 2021;24:835–43.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–80.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559.
Lin Y, Qi X, Chen J, Shen B. Multivariate competing endogenous RNA network characterization for cancer microRNA biomarker discovery: a novel bioinformatics model with application to prostate cancer metastasis. Precis Clin Med. 2022;5:pbac001.
Lin Y, Chen F, Shen L, Tang X, Du C, Sun Z, et al. Biomarker microRNAs for prostate cancer metastasis: screened with a network vulnerability analysis model. J Transl Med. 2018;16:134.
Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–84.
Huang H-Y, Lin Y-C-D, Cui S, Huang Y, Tang Y, Xu J, et al. miRTarBase update 2022: an informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022;50:D222–30.
Yan W, Liu X, Wang Y, Han S, Wang F, Liu X, et al. Identifying drug targets in pancreatic ductal adenocarcinoma through machine learning, analyzing biomolecular networks, and structural modeling. Front Pharmacol. 2020;11:534.
Csardi G, Nepusz T. The Igraph software package for complex network research. InterJournal. Complex Systems; 2006. p. 1695.
Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J Integr Biol. 2012;16:284–7.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–8.
Knox C, Wilson M, Klinger CM, Franklin M, Oler E, Wilson A, et al. DrugBank 6.0: the DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024;52:D1265–75.
Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–62.
Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630:493–500.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61.
Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–65.
Kim T, Croce CM. MicroRNA: trends in clinical trials of cancer diagnosis and therapy strategies. Exp Mol Med. 2023;55:1314–21.
Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics—challenges and potential solutions. Nat Rev Drug Discov. 2021;20:629–51.
Sun Z, Zhang Q, Yuan W, Li X, Chen C, Guo Y, et al. MiR-103a-3p promotes tumour glycolysis in colorectal cancer via hippo/YAP1/HIF1A axis. J Exp Clin Cancer Res. 2020;39:250.
Leslie PL, Franklin DA, Liu Y, Zhang Y. p53 regulates the expression of LRP1 and apoptosis through a stress intensity-dependent microRNA feedback loop. Cell Rep. 2018;24:1484–95.
Wang P, Zhou Y, Wang J, Zhou Y, Zhang X, Liu Y, et al. miR-107 reverses the multidrug resistance of gastric cancer by targeting the CGA/EGFR/GATA2 positive feedback circuit. J Biol Chem. 2024;300:107522.
Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7.
Xu W, Yang Z, Xie C, Zhu Y, Shu X, Zhang Z, et al. PTEN lipid phosphatase inactivation links the hippo and PI3K/Akt pathways to induce gastric tumorigenesis. J Exp Clin Cancer Res. 2018;37:198.
Messina B, Lo Sardo F, Scalera S, Memeo L, Colarossi C, Mare M, et al. Hippo pathway dysregulation in gastric cancer: from Helicobacter pylori infection to tumor promotion and progression. Cell Death Dis. 2023;14:21.
Baird RD, Tan DS, Kaye SB. Weekly paclitaxel in the treatment of recurrent ovarian cancer. Nat Rev Clin Oncol. 2010;7:575–82.
Yoon HH. Ramucirumab plus paclitaxel for gastric cancer in China. Lancet Gastroenterol Hepatol. 2021;6:975–6.
Acknowledgements
First of all, I would like to express my sincere gratitude to my supervisor, Prof. Weichang Chen, for your careful guidance and unwavering support, which greatly benefited me in the process of selecting the topic, designing the research methodology and writing the thesis. Your rigorous attitude and profound knowledge have provided endless insights into my research and pushed me to continuously strive for excellence. Meanwhile, I would like to thank my supervisors, Prof Yan Wenying and Prof Shi Tongguo, who have given me important guidance and support in my research journey, and whose high-level insights and rich research experience have enabled me to avoid many misunderstandings in my research and provided valuable support for my academic growth. During the sample collection process of this project, I am especially grateful to all the colleagues who participated in the sample collection work. Without their hard work and precision, this study could not have been carried out smoothly. Finally, I would like to thank all those who have helped and supported me throughout my academic career.
Funding
This research was supported by the Key Research and Development Programme of Jiangsu Province (BE2020656); Medical and Health Science and Technology Innovation Project of Suzhou (SKY2022010, SKYD2022097); Foundation of Suzhou Medical College of Soochow University (MP13405423, MX13401423); National Natural Science Foundation of China (82270561, 82073156); Jiangsu Provincial Medical Key Discipline (ZDXK202246); and the Priority Academic Programme Development of Jiangsu Higher Education Institutions (SKYD2022097).
Author information
Authors and Affiliations
Contributions
Jiachun Lu: writing—original draft, formal analysis, investigation, methodology, software, visualisation, data curation. Yuqi Chen: conceptualisation, methodology, investigation, resources. Jiayu Wang: conceptualisation, methodology, investigation. Yuxin He: methodology, investigation. Xin Liu: methodology, investigation. Tongguo Shi: project administration, methodology, supervision, validation, writing—review & editing. Weichang Chen: conceptualisation, funding acquisition, project administration, supervision, writing—review & editing. Wenying Yan: conceptualisation, methodology, project administration, supervision, funding acquisition, writing—review & editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institution and with the 1964 Helsinki Declaration. Written informed consent was obtained from individual or guardian participants. The study was approved by the Bioethics Committee of the First Affiliated Hospital of Soochow University (Approval No. 2021068). All animal experiments were performed in accordance with the institutional guidelines of the Soochow Animal Care and Use Committee (No.202412A0046).
Consent for publication
The work described has not been previously published, the consent to publish has been obtained from all co-authors, and the publication has been approved by the competent authorities of the institution in which the work was carried out.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, J., Chen, Y., Liu, X. et al. Artificial intelligence-driven microRNA signature for early detection of gastric cancer: discovery and clinical functional exploration. Br J Cancer 132, 957–972 (2025). https://doi.org/10.1038/s41416-025-02984-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-025-02984-9