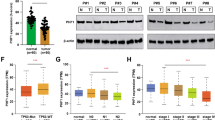
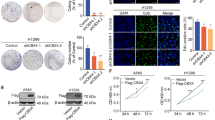
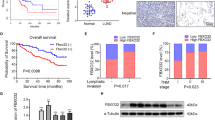
Abstract
Dysregulation of histone supply is implicated in various cancers, including lung adenocarcinoma (LUAD), although the underlying mechanisms remain poorly understood. Here, we demonstrate that knockout of Fbxo45 in mouse alveolar epithelial type 2 (AT2) cells leads to spontaneous LUAD. Our findings reveal that FBXO45 is a novel cell-cycle-regulated protein that is degraded upon phosphorylation by CDK1 during the S/G2 phase. During the S phase or DNA damage repair, FBXO45 binds to UPF1 and recruits the phosphatase PPP6C, thereby inhibiting UPF1 phosphorylation. This process is crucial for preventing the degradation of replication-dependent (RD) histone mRNAs and ensuring an adequate histone supply. In the absence of FBXO45, the impaired interaction between PPP6C and UPF1 results in sustained hyperphosphorylation of UPF1 throughout the cell cycle, leading to an insufficient histone supply, chromatin relaxation, genomic instability, and an increased rate of gene mutations, ultimately culminating in malignant transformation. Notably, analysis of clinical LUAD specimens confirms a positive correlation between the loss of FBXO45 and genomic instability, which is consistent with our findings in the mouse model. These results highlight the critical role of FBXO45 as a genomic guardian in coordinating histone supply and DNA replication, providing valuable insights into potential therapeutic targets and strategies for the treatment of LUAD.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The RNA-Seq data generated in this study were provided as EXCEL profiles in Supplementary Data and have been deposited in the Gene Expression Omnibus (GEO) repository under the accession number GSE218662. The whole-exome sequencing (WES) data generated in this study were provided as EXCEL profiles in Supplementary Data and have been deposited in the Sequence Read Archive (SRA) database under the accession number PRJNA905170. The mass spectrometry data generated in this study were provided as EXCEL profiles in Supplementary Data and have been uploaded to the Integrated Proteome Resources (iProX) database under the accession number IPX0005469000 and IPX0010103000. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All data are also available from the corresponding author (J.Y.) upon reasonable request.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49.
Chevallier M, Borgeaud M, Addeo A, Friedlaender A. Oncogenic driver mutations in non-small cell lung cancer: Past, present and future. World J Clin Oncol. 2021;12:217–37.
Nair N, Shoaib M, Sorensen CS. Chromatin dynamics in genome stability: roles in suppressing endogenous DNA damage and facilitating DNA repair. Int J Mol Sci. 2017;18:1486.
Zhao S, Allis CD, Wang GG. The language of chromatin modification in human cancers. Nat Rev Cancer. 2021;21:413–30.
O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol. 2010;17:1218–25.
Pal S, Tyler JK. Epigenetics and aging. Sci Adv. 2016;2:e1600584.
Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, et al. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–35.
Mejlvang J, Feng Y, Alabert C, Neelsen KJ, Jasencakova Z, Zhao X, et al. New histone supply regulates replication fork speed and PCNA unloading. J Cell Biol. 2014;204:29–43.
Singh RK, Liang D, Gajjalaiahvari UR, Kabbaj MH, Paik J, Gunjan A. Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle. 2010;9:4236–44.
Li L, Zhang X, Tian T, Pang L. Mathematical modelling the pathway of genomic instability in lung cancer. Sci Rep. 2019;9:14136.
Marzluff WF, Koreski KP. Birth and Death of Histone mRNAs. Trends Genet. 2017;33:745–59.
Davila Lopez M, Samuelsson T. Early evolution of histone mRNA 3’ end processing. RNA. 2008;14:1–10.
Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol. 2005;12:794–800.
Kaufman PD. Toxicity and lifespan extension: complex outcomes of histone overexpression in budding yeast. Cell Cycle. 2010;9:4611–2.
Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–54.
Meaux SA, Holmquist CE, Marzluff WF. Role of oligouridylation in normal metabolism and regulated degradation of mammalian histone mRNAs. Philos Trans R Soc Lond B Biol Sci. 2018;373:20180170.
Choe J, Ahn SH, Kim YK. The mRNP remodeling mediated by UPF1 promotes rapid degradation of replication-dependent histone mRNA. Nucleic Acids Res. 2014;42:9334–49.
Gowravaram M, Bonneau F, Kanaan J, Maciej VD, Fiorini F, Raj S, et al. A conserved structural element in the RNA helicase UPF1 regulates its catalytic activity in an isoform-specific manner. Nucleic Acids Res. 2018;46:2648–59.
Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, et al. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell. 2011;41:693–703.
Fiorini F, Bonneau F, Le Hir H. Biochemical characterization of the RNA helicase UPF1 involved in nonsense-mediated mRNA decay. Methods Enzymol. 2012;511:255–74.
Kim YK, Maquat LE. UP Front and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA. 2019;25:407–22.
Choe J, Kim KM, Park S, Lee YK, Song OK, Kim MK, et al. Rapid degradation of replication-dependent histone mRNAs largely occurs on mRNAs bound by nuclear cap-binding proteins 80 and 20. Nucleic Acids Res. 2013;41:1307–18.
Muller B, Blackburn J, Feijoo C, Zhao X, Smythe C. DNA-activated protein kinase functions in a newly observed S phase checkpoint that links histone mRNA abundance with DNA replication. J Cell Biol. 2007;179:1385–98.
Xu M, Zhu C, Zhao X, Chen C, Zhang H, Yuan H, et al. Atypical ubiquitin E3 ligase complex Skp1-Pam-Fbxo45 controls the core epithelial-to-mesenchymal transition-inducing transcription factors. Oncotarget. 2015;6:979–94.
Yoshida K. Characterization of estrogen-induced F-box protein FBXO45. Oncol Rep. 2005;14:531–5.
Yoon KA, Park JH, Han J, Park S, Lee GK, Han JY, et al. A genome-wide association study reveals susceptibility variants for non-small cell lung cancer in the Korean population. Hum Mol Genet. 2010;19:4948–54.
Tuominen R, Jonsson G, Enerback C, Appelqvist F, Olsson H, Ingvar C, et al. Investigation of a putative melanoma susceptibility locus at chromosome 3q29. Cancer Genet. 2014;207:70–4.
Wu C, Miao X, Huang L, Che X, Jiang G, Yu D, et al. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet. 2011;44:62–6.
Feng M, Ye X, Chen B, Zhang J, Lin M, Zhou H, et al. Detection of circulating genetically abnormal cells using 4-color fluorescence in situ hybridization for the early detection of lung cancer. J Cancer Res Clin Oncol. 2021;147:2397–405.
Katz RL, Zaidi TM, Pujara D, Shanbhag ND, Truong D, Patil S, et al. Identification of circulating tumor cells using 4-color fluorescence in situ hybridization: Validation of a noninvasive aid for ruling out lung cancer in patients with low-dose computed tomography-detected lung nodules. Cancer Cytopathol. 2020;128:553–62.
Willatt L, Cox J, Barber J, Cabanas ED, Collins A, Donnai D, et al. 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am J Hum Genet. 2005;77:154–60.
Glassford, M.R., R.H. Purcell, S. Pass, M.M. Murphy, G.J. Bassell, J.G. Mulle, et al., Caregiver perspectives on a Child’s diagnosis of 3q29 Deletion: “We Can’t Just Wish This Thing Away”. J Dev Behav Pediatr, 2022;43:e94–e102.
Saiga T, Fukuda T, Matsumoto M, Tada H, Okano HJ, Okano H, et al. Fbxo45 forms a novel ubiquitin ligase complex and is required for neuronal development. Mol Cell Biol. 2009;29:3529–43.
Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190–4.
Zheng HC, Takano Y. NNK-induced lung tumors: a review of animal model. J Oncol. 2011;2011:635379.
Wikenheiser KA, Vorbroker DK, Rice WR, Clark JC, Bachurski CJ, Oie HK, et al. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci USA. 1993;90:11029–33.
Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–98.
Sittman DB, Graves RA, Marzluff WF. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci USA. 1983;80:1849–53.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523.
Dou Y, Kalmykova S, Pashkova M, Oghbaie M, Jiang H, Molloy KR, et al. Affinity proteomic dissection of the human nuclear cap-binding complex interactome. Nucleic Acids Res. 2020;48:10456–69.
Hoefig KP, Heissmeyer V. Degradation of oligouridylated histone mRNAs: see UUUUU and goodbye. Wiley Interdiscip Rev RNA. 2014;5:577–89.
Hammond D, Zeng K, Espert A, Bastos RN, Baron RD, Gruneberg U, et al. Melanoma-associated mutations in protein phosphatase 6 cause chromosome instability and DNA damage owing to dysregulated Aurora-A. J Cell Sci. 2013;126:3429–40.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Maskin CR, Raman R, Houvras Y. PPP6C, a serine-threonine phosphatase, regulates melanocyte differentiation and contributes to melanoma tumorigenesis through modulation of MITF activity. Sci Rep. 2022;12:5573.
Sobajima T, Kowalczyk KM, Skylakakis S, Hayward D, Fulcher LJ, Neary C, et al. PP6 regulation of Aurora A-TPX2 limits NDC80 phosphorylation and mitotic spindle size. J Cell Biol. 2023;222:e202205117.
Chen X, Miragaia RJ, Natarajan KN, Teichmann SA. A rapid and robust method for single cell chromatin accessibility profiling. Nat Commun. 2018;9:5345.
Rizzo JM, Bard JE, Buck MJ. Standardized collection of MNase-seq experiments enables unbiased dataset comparisons. BMC Mol Biol. 2012;13:15.
Frew AJ, Johnstone RW, Bolden JE. Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer Lett. 2009;280:125–33.
Igaz N, Kovacs D, Razga Z, Konya Z, Boros IM, Kiricsi M. Modulating chromatin structure and DNA accessibility by deacetylase inhibition enhances the anti-cancer activity of silver nanoparticles. Colloids Surf B Biointerfaces. 2016;146:670–7.
Luzhna L, Kathiria P, Kovalchuk O. Micronuclei in genotoxicity assessment: from genetics to epigenetics and beyond. Front Genet. 2013;4:131.
Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–94.
Griesbach E, Schlackow M, Marzluff WF, Proudfoot NJ. Dual RNA 3’-end processing of H2A.X messenger RNA maintains DNA damage repair throughout the cell cycle. Nat Commun. 2021;12:359.
Hauer MH, Seeber A, Singh V, Thierry R, Sack R, Amitai A, et al. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat Struct Mol Biol. 2017;24:99–107.
Chang KW, Lin CE, Tu HF, Chung HY, Chen YF, Lin SC. Establishment of a p53 null murine oral carcinoma cell line and the identification of genetic alterations associated with this carcinoma. Int J Mol Sci. 2020;21:9354.
Esposito MR, Binatti A, Pantile M, Coppe A, Mazzocco K, Longo L, et al. Somatic mutations in specific and connected subpathways are associated with short neuroblastoma patients’ survival and indicate proteins targetable at onset of disease. Int J Cancer. 2018;143:2525–36.
Han ZF, Lin ST, Zhong M, Yu DJ. Correlations of UGT1A1 gene polymorphisms with onset and prognosis of non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2020;24:9973–80.
Preussner J, Zhong J, Sreenivasan K, Gunther S, Engleitner T, Kunne C, et al. Oncogenic amplification of zygotic dux factors in regenerating p53-deficient muscle stem cells defines a molecular cancer subtype. Cell Stem Cell. 2018;23:794–805 e4.
Jonckheere N, Van Seuningen I. Integrative analysis of the cancer genome atlas and cancer cell lines encyclopedia large-scale genomic databases: MUC4/MUC16/MUC20 signature is associated with poor survival in human carcinomas. J Transl Med. 2018;16:259.
Reiterova J, Tesar V. Autosomal dominant polycystic kidney disease: from pathophysiology of cystogenesis to advances in the treatment. Int J Mol Sci. 2022;23:3317.
Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–85 e18.
Martinez-Jimenez F, Muinos F, Sentis I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20:555–72.
Dhatchinamoorthy K, Colbert JD, Rock KL. Cancer immune evasion through loss of MHC class I antigen presentation. Front Immunol. 2021;12:636568.
He Y, Rozeboom L, Rivard CJ, Ellison K, Dziadziuszko R, Yu H, et al. MHC class II expression in lung cancer. Lung Cancer. 2017;112:75–80.
Johnson AM, Bullock BL, Neuwelt AJ, Poczobutt JM, Kaspar RE, Li HY, et al. Cancer cell-intrinsic expression of MHC Class II regulates the immune microenvironment and response to Anti-PD-1 therapy in lung adenocarcinoma. J Immunol. 2020;204:2295–307.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58.
Lin M, Wang ZW, Zhu X. FBXO45 is a potential therapeutic target for cancer therapy. Cell Death Discov. 2020;6:55.
Kogure N, Yokobori T, Ogata K, Altan B, Mochiki E, Ohno T, et al. Low Expression of FBXO45 Is Associated with Gastric Cancer Progression and Poor Prognosis. Anticancer Res. 2017;37:191–6.
Abshire CF, Carroll JL, Dragoi AM. FLASH protects ZEB1 from degradation and supports cancer cells’ epithelial-to-mesenchymal transition. Oncogenesis. 2016;5:e254.
Peschiaroli A, Scialpi F, Bernassola F, Pagano M, Melino G. The F-box protein FBXO45 promotes the proteasome-dependent degradation of p73. Oncogene. 2009;28:3157–66.
Chen X, Sahasrabuddhe AA, Szankasi P, Chung F, Basrur V, Rangnekar VM, et al. Fbxo45-mediated degradation of the tumor-suppressor Par-4 regulates cancer cell survival. Cell Death Differ. 2014;21:1535–45.
Richter KT, Kschonsak YT, Vodicska B, Hoffmann I. FBXO45-MYCBP2 regulates mitotic cell fate by targeting FBXW7 for degradation. Cell Death Differ. 2020;27:758–72.
Wu L, Yu K, Chen K, Zhu X, Yang Z, Wang Q, et al. Fbxo45 facilitates pancreatic carcinoma progression by targeting USP49 for ubiquitination and degradation. Cell Death Dis. 2022;13:231.
Wang Q, Wu L, Cao R, Gao J, Chai D, Qin Y, et al. Fbxo45 promotes the malignant development of esophageal squamous cell carcinoma by targeting GGNBP2 for ubiquitination and degradation. Oncogene. 2022;41:4795–807.
Al Zubaidi T, Gehrisch OHF, Genois MM, Liu Q, Lu S, Kung J, et al. Targeting the DNA replication stress phenotype of KRAS mutant cancer cells. Sci Rep. 2021;11:3656.
Chung FZ, Sahasrabuddhe AA, Ma K, Chen X, Basrur V, Lim MS, et al. Fbxo45 inhibits calcium-sensitive proteolysis of N-cadherin and promotes neuronal differentiation. J Biol Chem. 2014;289:28448–59.
Deem AK, Li X, Tyler JK. Epigenetic regulation of genomic integrity. Chromosoma. 2012;121:131–51.
Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–7.
Gionchiglia N, Granato A, Merighi A, Lossi L. Association of Caspase 3 Activation and H2AX gamma Phosphorylation in the Aging Brain: Studies on Untreated and Irradiated Mice. Biomedicines. 2021;9:1166.
Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair. 2004;3:959–67.
Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–9.
Sinha M, Lowell CA. Isolation of highly pure primary mouse alveolar epithelial type II cells by flow cytometric cell sorting. Bio Protoc. 2016;6:e2013.
Zhang H, Zhao X, Guo Y, Chen R, He J, Li L, et al. Hypoxia regulates overall mRNA homeostasis by inducing Met(1)-linked linear ubiquitination of AGO2 in cancer cells. Nat Commun. 2021;12:5416.
De Clerck NM, Meurrens K, Weiler H, Van Dyck D, Van G, Houtte, et al. High-resolution X-ray microtomography for the detection of lung tumors in living mice. Neoplasia. 2004;6:374–9.
Dzierzak E, de Bruijn M. Isolation and analysis of hematopoietic stem cells from mouse embryos. Methods Mol Med. 2002;63:1–14.
Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8.
Gansauge MT, Meyer M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat Protoc. 2013;8:737–48.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20.
Soederberg A, Meissgeier T, Bosserhoff AK, Linck-Paulus L. MAGOH and MAGOHB Knockdown in Melanoma Cells Decreases Nonsense-Mediated Decay Activity and Promotes Apoptosis via Upregulation of GADD45A. Cells. 2022;11:3859.
Baird TD, Cheng KC, Chen YC, Buehler E, Martin SE, Inglese J, et al. ICE1 promotes the link between splicing and nonsense-mediated mRNA decay. Elife. 2018;7:e33178.
Pereverzev AP, Gurskaya NG, Ermakova GV, Kudryavtseva EI, Markina NM, Kotlobay AA, et al. Method for quantitative analysis of nonsense-mediated mRNA decay at the single cell level. Sci Rep. 2015;5:7729.
Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–57.
Acknowledgements
We thank the Proteomics, Electron microscope and Flow sorting laboratory of Core Facility, Shanghai Jiao Tong University College of Basic Medical Sciences.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82230100, 32271310, 82273138) and Natural Science Foundation of Shanghai (23ZR1411500).
Author information
Authors and Affiliations
Contributions
JY and LL conceptualized the project; LL, JL, RC, CH, YZ and JC designed the methods; LL, JL, RC, CH, RL, XL, and JH performed most of experiments; LL, YW, XZ and JY performed formal analyses; LL and JY wrote the manuscript; JY, CD, and XJZ supervised the project. All authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Animal experiments were conducted following the general rules for the care and use of experimental animals and approved by the Shanghai Jiao Tong University School of Medicine Animal Care and Use Committee (permission no. A-2022-036). Clinical samples were collected from patients with written informed consent, following a protocol approved by The Affiliated Renji Hospital of Shanghai Jiao Tong University School of Medicine (ethics approval no. RA-2022-267).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, L., Li, J., Chen, R. et al. Loss of Fbxo45 in AT2 cells leads to insufficient histone supply and initiates lung adenocarcinoma. Cell Death Differ (2024). https://doi.org/10.1038/s41418-024-01433-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41418-024-01433-z