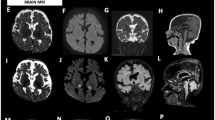
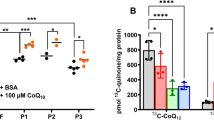
Abstract
Primary Coenzyme Q10 (CoQ10) deficiencies are a group of clinically heterogenous mitochondrial disorders that result from defects in CoQ10 biosynthesis. Their diagnosis is complicated by the absence of pathognomonic signs and poor genotype-phenotype correlations. Pathogenic variants in the COQ9 gene are a rare cause of CoQ10 deficiency: few cases have been reported, and the clinical presentation was described as a very severe multisystemic disorder with neonatal onset, ultimately leading to premature death. Through exome sequencing, we identified a novel homozygous splicing variant c.73 G > A in the COQ9 gene (NG_027696.1, NM_020312.4) in two adult siblings who presented with pure spastic paraplegia with onset in childhood. mRNA analysis from different tissues of one of the siblings revealed that this variant alters COQ9 splicing, resulting in undetectable levels of COQ9 and COQ7 proteins and reduced concentrations of CoQ10 in muscle and fibroblasts. Additionally, the accumulation of 6-demethoxycoenzyme Q10, the substrate of COQ7, was observed in both plasma and fibroblasts. Furthermore, fibroblast proliferation rate was reduced when enhancing the mitochondrial metabolism by replacing glucose with galactose in the culture medium, and was rescued by the addition of exogenous CoQ10, suggesting a therapeutic avenue for these patients. Altogether, we report here the first example of hereditary spastic paraplegia related to a mutation of the COQ9 gene that expands the spectrum of clinical manifestations and opens new therapeutic opportunities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
References
Turunen M, Olsson J, Dallner G. Metabolism and function of Coenzyme Q. Biochim Biophys Acta. 2004;1660:171–199. https://doi.org/10.1016/j.bbamem.2003.11.012.
Guerra RM, Pagliarini DJ. Coenzyme Q biochemistry and biosynthesis. Trends Biochem Sci. 2023;48:463–476. https://doi.org/10.1016/j.tibs.2022.12.006.
Mantle D, Millichap L, Castro-Marrero J, Hargreaves IP. Primary coenzyme Q10 deficiency: an update. Antioxidants. 2023;12:1652.
Salviati L, Trevisson E, Agosto C, Doimo M, Navas P (2017). Primary coenzyme Q10 deficiency. In GeneReviews® 2017.
Paprocka J, Nowak M, Chuchra P, Śmigiel R. COQ8A-Ataxia as a manifestation of primary Coenzyme Q deficiency. Metabolites. 2022;12:955 https://doi.org/10.3390/metabo12100955.
Drovandi S, Lipska-Ziętkiewicz BS, Ozaltin F, Emma F, Gulhan B, Boyer O, et al. Variation of the clinical spectrum and genotype-phenotype associations in Coenzyme Q10 deficiency-associated glomerulopathy. Kidney Int. 2022;102:592–603. https://doi.org/10.1016/j.kint.2022.02.040.
Jacquier A, Theuriet J, Fontaine F, Mosbach V, Lacoste N, Ribault S, et al. Homozygous COQ7 mutation: a new cause of potentially treatable distal hereditary motor neuropathy. Brain. 2023;146:3470–3483. https://doi.org/10.1093/brain/awac453.
Wang Y, Hekimi S. The efficacy of coenzyme Q10 treatment in alleviating the symptoms of primary coenzyme Q10 deficiency: A systematic review. J Cell Mol Med. 2022;26:4635–4644. https://doi.org/10.1111/jcmm.17488.
Manicki M, Aydin H, Abriata LA, Overmyer KA, Guerra RM, Coon JJ, et al. Structure and functionality of a multimeric human COQ7:COQ9 complex. Mol Cell. 2022;82:4307–4323. https://doi.org/10.1016/j.molcel.2022.10.003.
Duncan AJ, Bitner-Glindzicz M, Meunier B, Costello H, Hargreaves IP, López LC, et al. A nonsense mutation in COQ9 causes autosomal-recessive neonatal-onset primary Coenzyme Q10 deficiency: a potentially treatable form of mitochondrial disease. Am J Hum Genet. 2009;84:558–566. https://doi.org/10.1016/j.ajhg.2009.03.018.
Danhauser K, Herebian D, Haack TB, Rodenburg RJ, Strom TM, Meitinger T, et al. Fatal neonatal encephalopathy and lactic acidosis caused by a homozygous loss-of-function variant in COQ9. Eur J Hum Genet. 2016;24:450–454. https://doi.org/10.1038/ejhg.2015.133.
Smith AC, Ito Y, Ahmed A, Schwartzentruber JA, Beaulieu CL, Aberg E, et al. A family segregating lethal neonatal Coenzyme Q10 deficiency caused by mutations in COQ9. J Inherit Metab Dis. 2018;41:719–729. https://doi.org/10.1007/s10545-017-0122-7.
Olgac A, Oztoprak U, Kasapkara CS, Kılıç M, Yüksel D, Derinkuyu EB, et al. A rare case of primary Coenzyme Q10 deficiency due to COQ9 mutation. J Pediatr Endocrinol Metab. 2020;33:165–170. https://doi.org/10.1515/jpem-2019-0245.
Dimassi S, Simonet T, Labalme A, Boutry-Kryza N, Campan-Fournier A, Lamy R, et al. Comparison of two next-generation sequencing kits for diagnosis of epileptic disorders with a user-friendly tool for displaying gene coverage, DeCovA. Appl Transl Genom. 2015;17:19–25. https://doi.org/10.1016/j.atg.2015.10.001.
Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016;11:1–9. https://doi.org/10.1038/nprot.2015.123.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. https://doi.org/10.1038/nmeth0410-248.
Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. https://doi.org/10.1038/nmeth.2890.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. https://doi.org/10.1038/ng.2892.
Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: An ensemble method for predicting the pathogenicity of rare Missense variants. Am J Hum Genet. 2016;99:877–885. https://doi.org/10.1016/j.ajhg.2016.08.016.
Tordai H, Torres O, Csepi M, Padányi R, Lukács GL, Hegedűs T. Analysis of AlphaMissense data in different protein groups and structural context. Sci Data. 2024;11:495. https://doi.org/10.1038/s41597-024-03327-8.
Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–52. https://doi.org/10.1093/nar/gkr485.
Schluth-Bolard C, Diguet F, Chatron N, Rollat-Farnier PA, Bardel C, Afenjar A, et al. Whole genome paired-end sequencing elucidates functional and phenotypic consequences of balanced chromosomal rearrangement in patients with developmental disorders. J Med Genet. 2019;56:526–535. https://doi.org/10.1136/jmedgenet-2018-105778.
Medja F, Allouche S, Frachon P, Jardel C, Malgat M, Mousson de Camaret B, et al. Development and implementation of standardized respiratory chain spectrophotometric assays for clinical diagnosis. Mitochondrion. 2009;9:331–339. https://doi.org/10.1016/j.mito.2009.05.001.
Fontaine F, Legallois D, Créveuil C, Chtourou M, Coulbault L, Milliez P, et al. Is plasma concentration of coenzyme Q10 a predictive marker for left ventricular remodelling after revascularization for ST-segment elevation myocardial infarction?. Ann Clin Biochem. 2021;58:327–334. https://doi.org/10.1177/00045632211001100.
Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, et al. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. https://doi.org/10.1016/0003-2697(85)90442-7.
Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–985. https://doi.org/10.1093/nar/gkt1113.
Fokkema IF, Kroon M, López Hernández JA, Asscheman D, Lugtenburg I, Hoogenboom J, et al. The LOVD3 platform: efficient genome-wide sharing of genetic variants. Eur J Hum Genet. 2021;29:1796–1803. https://doi.org/10.1038/s41431-021-00959-x.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. https://doi.org/10.1038/s41586-020-2308-7.
Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R, et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35:1978–1980. https://doi.org/10.1093/bioinformatics/bty897.
Baux D, Van Goethem C, Ardouin O, Guignard T, Bergougnoux A, Koenig M, et al. MobiDetails: online DNA variants interpretation. Eur J Hum Genet. 2021;29:356–360.
Leman R, Parfait B, Vidaud D, Girodon E, Pacot L, Le Gac G, et al. SPiP: Splicing Prediction Pipeline, a machine learning tool for massive detection of exonic and intronic variant effects on mRNA splicing. Hum Mutat. 2022;43:2308–2323. https://doi.org/10.1002/humu.24491.
Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: A one-stop database of functional predictions and annotations for human nonsynonymous and Splice-Site SNVs. Hum Mutat. 2016;37:235–41. https://doi.org/10.1002/humu.22932.
Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24. https://doi.org/10.1016/j.cell.2018.12.015.
Yubero D, Allen G, Artuch R, Montero R. The value of Coenzyme Q10 determination in mitochondrial patients. J Clin Med. 2017;6:37–47. https://doi.org/10.3390/jcm6040037.
Herebian D, Seibt A, Smits SHJ, Bünning G, Freyer C, Prokisch H, et al. Detection of 6-demethoxyubiquinone in CoQ10 deficiency disorders: insights into enzyme interactions and identification of potential therapeutics. Mol Genet Metab. 2017;121:216–223. https://doi.org/10.1016/j.ymgme.2017.05.012.
Alcázar-Fabra M, Rodríguez-Sánchez F, Trevisson E, Brea-Calvo G. Primary Coenzyme Q deficiencies: a literature review and online platform of clinical features to uncover genotype-phenotype correlations. Free Radic Biol Med. 2021;167:141–180. https://doi.org/10.1016/j.freeradbiomed.2021.02.046.
Luna-Sánchez M, Díaz-Casado E, Barca E, Tejada MA, Montilla-García A, Cobos EJ, et al. The clinical heterogeneity of coenzyme Q10 deficiency results from genotypic differences in the Coq9 gene. EMBO Mol Med. 2015;7:670–687. https://doi.org/10.15252/emmm.201404632.
Wongkittichote P, Duque Lasio ML, Magistrati M, Pathak S, Sample B, Rocha Carvalho D, et al. Phenotypic, molecular, and functional characterization of COQ7-related primary CoQ10 deficiency: hypomorphic variants and two distinct disease entities. Mol Genet Metab. 2023;139:107630. https://doi.org/10.1016/j.ymgme.2023.107630.
Wang Y, Smith C, Parboosingh JS, Khan A, Innes M, Hekimi S. Pathogenicity of two COQ7 mutations and responses to 2,4-dihydroxybenzoate bypass treatment. J Cell Mol Med. 2017;21:2329–2343. https://doi.org/10.1111/jcmm.13154.
Freyer C, Stranneheim H, Naess K, Mourier A, Felser A, Maffezzini C, et al. Rescue of primary ubiquinone deficiency due to a novel COQ7 defect using 2,4-dihydroxybensoic acid. J Med Genet. 2015;52:779–783. https://doi.org/10.1136/jmedgenet-2015-102986.
López-Martín JM, Salviati L, Trevisson E, Montini G, DiMauro S, Quinzii C, et al. Missense mutation of the COQ2 gene causes defects of bioenergetics and de novo pyrimidine synthesis. Hum Mol Genet. 2007;16:1091–1097. https://doi.org/10.1093/hmg/ddm058.
Quinzii CM, Luna-Sanchez M, Ziosi M, Hidalgo-Gutierrez A, Kleiner G, Lopez LC. The role of sulfide oxidation impairment in the pathogenesis of primary CoQ deficiency. Front Physiol. 2017;8:525 https://doi.org/10.3389/fphys.2017.00525.
Rodríguez-Hernández A, Cordero MD, Salviati L, Artuch R, Pineda M, Briones P, et al. Coenzyme Q deficiency triggers mitochondria degradation by mitophagy. Autophagy. 2009;5:19–32. https://doi.org/10.4161/auto.5.1.7174.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
Writing – original draft, F.F.; writing – review & editing, S.A., G.L., F.F., A.J., J.T.; conceptualization, G.L., S.A., F.F., A.J., J.T.; formal analysis, S.A., A.J.; methodology, G.L., S.A., F.F., A.J., J.T.; investigation – in vitro and variant analysis, F.F., A.L., J.T., A.J., N.L., N.S.; investigation – clinical, C.L.; visualization, F.F., A.J.; supervision, S.A., G.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All procedures performed in this study involving human participants were carried out in accordance with the ethical standards of the University Hospital of Caen, and the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Informed consent was obtained from both patients included in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fontaine, F., Labalme, A., Laurencin, C. et al. Homozygous COQ9 mutation: a new cause of potentially treatable hereditary spastic paraplegia. Eur J Hum Genet (2025). https://doi.org/10.1038/s41431-025-01895-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41431-025-01895-w