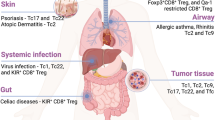
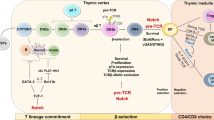
Abstract
CD8+ T cells play a critical role in specific immunity. In recent years, cell therapy has been emerging rapidly. The specific cytotoxic capabilities of these cells enable them to precisely identify and kill cells presenting specific antigens. This has demonstrated promise in the treatment of autoimmune diseases and cancers, with wide-ranging applications and value. However, in some diseases, such as tumors and chronic infections, T cells may adopt an exhausted phenotype, resulting in a loss of cytotoxicity and limiting their further application. Epigenetics plays a significant role in the differentiation and regulation of gene expression in cells. There is extensive evidence indicating that epigenetic remodeling plays an important role in T cell exhaustion. Therefore, further understanding its role in CD8+ T cell function can provide insights into the programmatic regulation of CD8+ T cells from a genetic perspective and overcome these diseases. We attempted to describe the relationship between the activation, function, and exhaustion mechanisms of CD8+ T cells, as well as epigenetics. This understanding makes it possible for us to address the aforementioned issues.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
118,99 € per year
only 19,83 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ma X, Xiao L, Liu L, Ye L, Su P, Bi E, et al. CD36-mediated ferroptosis dampens intratumoral CD8+ T cell effector function and impairs their antitumor ability. Cell Metab. 2021;33:1001.
Rudloff MW, Zumbo P, Favret NR, Roetman JJ, Detrés Román CR, Erwin MM, et al. Hallmarks of CD8+ T cell dysfunction are established within hours of tumor antigen encounter before cell division. Nat Immunol. 2023;24:1527–39.
Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–9.
Huang Y, Si X, Shao M, Teng X, Xiao G, Huang H. Rewiring mitochondrial metabolism to counteract exhaustion of CAR-T cells. J Hematol Oncol. 2022;15:38.
Beltra J-C, Manne S, Abdel-Hakeem MS, Kurachi M, Giles JR, Chen Z, et al. Developmental Relationships of Four Exhausted CD8+ T Cell Subsets Reveals Underlying Transcriptional and Epigenetic Landscape Control Mechanisms. Immunity. 2020;52:825.
Belk JA, Daniel B, Satpathy AT. Epigenetic regulation of T cell exhaustion. Nat Immunol. 2022;23:848–60.
McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. In: Yokoyama WM, editor. Annu Rev Immunol. 2019;37:457–95. Annual Review of Immunology. 372019
Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–99.
Hu Y, Zhou Y, Zhang M, Ge W, Li Y, Yang L, et al. CRISPR/Cas9-Engineered Universal CD19/CD22 Dual-Targeted CAR-T Cell Therapy for Relapsed/Refractory B-cell Acute Lymphoblastic Leukemia. Clin Cancer Res. 2021;27:2764–72.
Ledford H. CAR-T Therapy Forces Autoimmune Diseases into Remission. Nature. 2023;624:483–4.
Arnold C. Autoimmune disease is the next frontier for CAR T cell therapy. Nat Med. 2024;30:6–9.
Horste EL, Fansler MM, Cai T, Chen X, Mitschka S, Zhen G, et al. Subcytoplasmic ___location of translation controls protein output. Mol cell. 2023;83:4509–23.e11.
Wang Z, Wu Z, Liu Y, Han W. New development in CAR-T cell therapy. J Hematol Oncol. 2017;10:53.
Papp F, Hajdu P, Tajti G. et al. Periodic Membrane Potential and Ca2+ Oscillations in T Cells Forming an Immune Synapse. Int J Mol Sci. 2020;21:1568.
Green DR, Droin N, Pinkoski M. Activation-induced cell death in T cells. Immunological Rev. 2003;193:70–81.
Denton AE, Wesselingh R, Gras S, Guillonneau C, Olson MR, Mintern JD, et al. Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses. J Immunol. 2011;187:5733–44.
Duan H, Jing L, Jiang X, Ma Y, Wang D, Xiang J. et al. CD146 bound to LCK promotes T cell receptor signaling and antitumor immune responses in mice. J Clin Invest. 2021;131:e148568
Kasmani MY, Zander R, Chung HK. et al. Clonal lineage tracing reveals mechanisms skewing CD8+ T cell fate decisions in chronic infection. J Exp Med. 2023;220:e20220679.
Beltra JC, Manne S, Abdel-Hakeem MS, Kurachi M, Giles JR, Chen Z, et al. Developmental Relationships of Four Exhausted CD8(+) T Cell Subsets Reveals Underlying Transcriptional and Epigenetic Landscape Control Mechanisms. Immunity. 2020;52:825–41.e8.
Blank CU, Haining WN, Held W, Hogan PG, Kallies A, Lugli E, et al. Defining ‘T cell exhaustion. Nat Rev Immunol. 2019;19:665–74.
Schietinger A, Philip M, Krisnawan VE, Chiu EY, Delrow JJ, Basom RS, et al. Tumor-Specific T Cell Dysfunction Is a Dynamic Antigen-Driven Differentiation Program Initiated Early during Tumorigenesis. Immunity. 2016;45:389–401.
Siddiqui I, Schaeuble K, Chennupati V, Marraco SAF, Calderon-Copete S, Ferreira DP, et al. Intratumoral Tcf1 + PD-1 + CD8 + T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity. 2019;50:195.
Ozga AJ, Chow MT, Lopes ME, Servis RL, Di Pilato M, Dehio P, et al. CXCL10 chemokine regulates heterogeneity of the CD8 + T cell response and viral set point during chronic infection. Immunity. 2022;55:82.
Pritykin Y, van der Veeken J, Pine AR, Zhong Y, Sahin M, Mazutis L, et al. A unified atlas of CD8 T cell dysfunctional states in cancer and infection. Mol Cell. 2021;81:2477.
McKinney EF, Lee JC, Jayne DRW, Lyons PA, Smith KGC. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 2015;523:612.
Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell. 2019;176:775.
Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8 + T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20:326.
Shan Q, Zeng Z, Xing S, Li F, Hartwig SM, Gullicksrud JA, et al. The transcription factor Runx3 guards cytotoxic CD8 + effector T cells against deviation towards follicular helper T cell lineage. Nat Immunol. 2017;18:931.
Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, et al. CXCR5 + follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol. 2016;17:1187.
Weng NP, Akbar AN, Goronzy J. CD28(-) T cells: their role in the age-associated decline of immune function. Trends Immunol. 2009;30:306–12.
Seyda M, Elkhal A, Quante M, Falk CS, Tullius SG. T Cells Going Innate. Trends Immunol. 2016;37:546–56.
Chen X, Liu Q, Xiang AP. CD8+CD28- T cells: not only age-related cells but a subset of regulatory T cells. Cell Mol Immunol. 2018;15:734–6.
Larbi A, Pawelec G, Wong SC, Goldeck D, Tai JJ, Fulop T. Impact of age on T cell signaling: a general defect or specific alterations? Ageing Res Rev. 2011;10:370–8.
Huff WX, Kwon JH, Henriquez M, Fetcko K, Dey M. The Evolving Role of CD8+CD28− Immunosenescent T Cells in Cancer Immunology. Int J Mol Sci. 2019;20:2810.
Dai D, Pei Y, Zhu B, Wang D, Pei S, Huang H, et al. Chemoradiotherapy-induced ACKR2+ tumor cells drive CD8+ T cell senescence and cervical cancer recurrence. Cell Rep Med. 2024;5:101550.
Guan Y, Zhang C, Lyu G, Huang X, Zhang X, Zhuang T, et al. Senescence-activated enhancer landscape orchestrates the senescence-associated secretory phenotype in murine fibroblasts. Nucleic Acids Res. 2020;48:10909–23.
Pomp W, Meeussen JVW, Lenstra TL. Transcription factor exchange enables prolonged transcriptional bursts. Mol Cell. 2024;84:1036–1048.e9.
Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–402.
Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and Function of Mammalian DNA Methyltransferases. Chembiochem. 2011;12:206–22.
Ghanty U, Wang T, Kohli RM. Nucleobase Modifiers Identify TET Enzymes as Bifunctional DNA Dioxygenases Capable of Direct N-Demethylation. Angew Chem-Int Ed. 2020;59:11312–5.
DNMT3A DNA-Binding Residues Provide Specificity for CpG DNA Methylation. Cancer discovery. 2018;8:OF14-OF.
Zhang Z-M, Lu R, Wang P, Yu Y, Chen D, Gao L, et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature. 2018;554:387.
Nunez JK, Chen J, Pommier GC, Cogan JZ, Replogle JM, Adriaens C, et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell. 2021;184:2503.
Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–57.
Kschonsak M, Haering CH. Shaping mitotic chromosomes: From classical concepts to molecular mechanisms. Bioessays. 2015;37:755–66.
Eustermann S, Patel AB, Hopfner K-P, He Y, Korber P. Energy-driven genome regulation by ATP-dependent chromatin remodellers. Nat Rev Mol Cell Biol. 2023;25:309–32.
Song J, Gooding AR, Hemphill WO, Kasinath V, Cech TR. Structural basis for inactivation of PRC2 by G-quadruplex RNA. Science. 2023;381:1331–7.
Skvortsova K, Iovino N, Bogdanovic O. Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol. 2018;19:774–90.
Roundtree IA, Luo G-Z, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife. 2017;6:e31311.
Fitz-James MH, Cavalli G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat Rev Genet. 2022;23:325–41.
Jain N, Zhao Z, Koche RP, Antelope C, Gozlan Y, Montalbano A, et al. Disruption of SUV39H1-Mediated H3K9 Methylation Sustains CAR T-cell Function. Cancer Discov. 2024;14:142–57.
Gattinoni L, Klebanoff CA, Palmer DC, Wrzesinski C, Kerstann K, Yu ZY, et al. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8 + T cells. J Clin Investig. 2005;115:1616–26.
Pantaleo G, Harari A. Functional signatures in antiviral T-cell immunity for monitoring virus-associated diseases. Nat Rev Immunol. 2006;6:417–23.
Baessler A, Vignali DAA. T Cell Exhaustion. Annu Rev Immunol. 2024;42:179–206.
Yu B, Zhang K, Milner JJ, Toma C, Chen R, Scott-Browne JP, et al. Epigenetic landscapes reveal transcription factors that regulate CD8(+) T cell differentiation. Nat Immunol. 2017;18:573–82.
Yang R, Mele F, Worley L, Langlais D, Rosain J, Benhsaien I, et al. Human T-bet Governs Innate and Innate-like Adaptive IFN-gamma Immunity against Mycobacteria. Cell. 2020;183:1826–47 e31.
Zhao X, Shan Q, Xue H-H. TCF1 in T cell immunity: a broadened frontier. Nat Rev Immunol. 2021;22:147–57.
Gattinoni L, Zhong X-S, Palmer DC, Ji Y, Hinrichs CS, Yu Z, et al. Wnt signaling arrests effector T cell differentiation and generates CD8 + memory stem cells. Nat Med. 2009;15:808–U129.
Sun Q, Cai D, Liu D, Zhao X, Li R, Xu W, et al. BCL6 promotes a stem-like CD8(+) T cell program in cancer via antagonizing BLIMP1. Sci Immunol. 2023;8:eadh1306.
Jung IY, Narayan V, McDonald S, Rech AJ, Bartoszek R, Hong G, et al. BLIMP1 and NR4A3 transcription factors reciprocally regulate antitumor CAR T cell stemness and exhaustion. Sci Transl Med. 2022;14:eabn7336.
Li Q, Zhang L, You W, Xu J, Dai J, Hua D, et al. PRDM1/BLIMP1 induces cancer immune evasion by modulating the USP22-SPI1-PD-L1 axis in hepatocellular carcinoma cells. Nat Commun. 2022;13:7677.
Stairiker CJ, Thomas GD, Salek-Ardakani S. EZH2 as a Regulator of CD8+ T Cell Fate and Function. Front Immunol. 2020;11:593203.
Abadie K, Clark EC, Valanparambil RM, Ukogu O, Yang W, Daza RM, et al. Reversible, tunable epigenetic silencing of TCF1 generates flexibility in the T cell memory decision. Immunity. 2024;57:271–86.e13.
Wang Y, Qiu F, Xu Y, Hou X, Zhang Z, Huang L, et al. Stem cell-like memory T cells: The generation and application. J Leukoc Biol. 2021;110:1209–23.
Seo H, Chen J, González-Avalos E, Samaniego-Castruita D, Das A, Wang YH, et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8(+) T cell exhaustion. Proc Natl Acad Sci USA. 2019;116:12410–5.
Ferreira DP, Silva JG, Wyss T, Marraco SAF, Scarpellino L, Charmoy M, et al. Central memory CD8 + T cells derive from stem-like Tcf7 hi effector cells in the absence of cytotoxic differentiation. Immunity. 2020;53:985.
Oestreich KJ, Yoon H, Ahmed R, Boss JM. NFATc1 regulates PD-1 expression upon T cell activation. J Immunol. 2008;181:4832–9.
Doan AE, Mueller KP, Chen AY, Rouin GT, Chen Y, Daniel B, et al. FOXO1 is a master regulator of memory programming in CAR T cells. Nature. 2024;629:211–8.
McCutcheon SR, Swartz AM, Brown MC, Barrera A, McRoberts Amador C, Siklenka K, et al. Transcriptional and epigenetic regulators of human CD8(+) T cell function identified through orthogonal CRISPR screens. Nat Genet. 2023;55:2211–23.
Yang R, Mele F, Worley L, Langlais D, Rosain J, Benhsaien I, et al. Human T-bet Governs Innate and Innate-like Adaptive IFN-γ Immunity against Mycobacteria. Cell. 2020;183:1826–47.e31.
Hertweck A, Evans CM, Eskandarpour M, Lau JC, Oleinika K, Jackson I, et al. T-bet Activates Th1 Genes through Mediator and the Super Elongation Complex. Cell Rep. 2016;15:2756–70.
Li Y, Han M, Wei H, Huang W, Chen Z, Zhang T, et al. Id2 epigenetically controls CD8(+) T-cell exhaustion by disrupting the assembly of the Tcf3-LSD1 complex. Cell Mol Immunol. 2024;21:292–308.
Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell. 2013;153:307–19.
Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013;155:934–47.
Danilo M, Chennupati V, Silva JG, Siegert S, Held W. Suppression of Tcf1 by Inflammatory Cytokines Facilitates Effector CD8 T Cell Differentiation. Cell Rep. 2018;22:2107–17.
Zhou X, Yu S, Zhao D-M, Harty JT, Badovinac VP, Xue H-H. Differentiation and Persistence of Memory CD8 + T Cells Depend on T Cell Factor 1. Immunity. 2010;33:229–40.
Miller CH, Klawon DEJ, Zeng S, Lee V, Socci ND, Savage PA. Eomes identifies thymic precursors of self-specific memory-phenotype CD8 + T cells. Nat Immunol. 2020;21:567.
Llao-Cid L, Roessner PM, Chapaprieta V, Oeztuerk S, Roider T, Bordas M, et al. EOMES is essential for antitumor activity of CD8+T cells in chronic lymphocytic leukemia. Leukemia. 2021;35:3152–62.
Beltra J-C, Abdel-Hakeem MS, Manne S, Zhang Z, Huang H, Kurachi M, et al. Stat5 opposes the transcription factor Tox and rewires exhausted CD8+ Tcells toward durable effector-like states during chronic antigen exposure. Immunity. 2023;56:2699–718.e11.
Youngblood B, Hale JS, Kissick HT, Ahn E, Xu X, Wieland A, et al. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature. 2017;552:404–9.
Herek TA, Bouska A, Lone W, Sharma S, Amador C, Heavican TB, et al. DNMT3A mutations define a unique biological and prognostic subgroup associated with cytotoxic T cells in PTCL-NOS. Blood. 2022;140:1278–90.
Prinzing B, Zebley CC, Petersen CT, Fan Y, Anido AA, Yi Z, et al. Deleting DNMT3A in CAR T cells prevents exhaustion and enhances antitumor activity. Sci Transl Med. 2021;13:eabh0272.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009;324:930–5.
Dixon G, Pan H, Yang D, Rosen BP, Jashari T, Verma N, et al. QSER1 protects DNA methylation valleys from de novo methylation. Science. 2021;372:eabd0875.
Bamezai S, Demir D, Pulikkottil AJ, Ciccarone F, Fischbein E, Sinha A, et al. TET1 promotes growth of T-cell acute lymphoblastic leukemia and can be antagonized via PARP inhibition. Leukemia. 2021;35:389–403.
Whiteside SK, Grant FM, Alvisi G, Clarke J, Tang L, Imianowski CJ, et al. Acquisition of suppressive function by conventional T cells limits antitumor immunity upon Treg depletion. Sci Immunol. 2023;8:eabo5558.– eabo
Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An Interleukin-21-Interleukin-10-STAT3 Pathway Is Critical for Functional Maturation of Memory CD8 + T Cells. Immunity. 2011;35:792–805.
Sun Q, Zhao X, Li R, Liu D, Pan B, Xie B, et al. STAT3 regulates CD8 + T cell differentiation and functions in cancer and acute infection. J Exp Med. 2023;220:e20220686.
Klysz DD, Fowler C, Malipatlolla M, Stuani L, Freitas KA, Chen Y, et al. Inosine induces stemness features in CAR-T cells and enhances potency. Cancer Cell. 2024;42:266–82 e8.
Rodriguez PC, Ernstoff MS, Hernandez C, Atkins M, Zabaleta J, Sierra R, et al. Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer Res. 2009;69:1553–60.
Speiser DE, Ho PC, Verdeil G. Regulatory circuits of T cell function in cancer. Nat Rev Immunol. 2016;16:599–611.
Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell. 2015;162:1229–41.
Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52.
Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22:266–82.
Yu P, Zhang X, Liu N, Tang L, Peng C, Chen X. Pyroptosis: mechanisms and diseases. Sig Transduct Targeted Ther. 2021;6:128.
Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254.
Liu X, Nie L, Zhang Y, Yan Y, Wang C, Colic M, et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol. 2023;25:404.
Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–61.
Xiao L, Ma X, Ye L, Su P, Xiong W, Bi E, et al. IL-9/STAT3/fatty acid oxidation-mediated lipid peroxidation contributes to Tc9 cell longevity and enhanced antitumor activity. Journal of Clinical Investigation. 2022;132:e153247.
Vignali PDA, DePeaux K, Watson MJJ, Ye C, Ford BR, Lontos K, et al. Hypoxia drives CD39-dependent suppressor function in exhausted T cells to limit antitumor immunity. Nat Immunol. 2023;24:267.
Alizadeh D, Wong RA, Yang X, Wang D, Pecoraro JR, Kuo CF, et al. IL15 Enhances CAR-T Cell Antitumor Activity by Reducing mTORC1 Activity and Preserving Their Stem Cell Memory Phenotype. Cancer Immunol Res. 2019;7:759–72.
MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–83.
Wang F, Cheng F, Zheng F. Stem cell like memory T cells: A new paradigm in cancer immunotherapy. Clin Immunol. 2022;241:109078.
Chang CH, Curtis JD, Maggi LB Jr, Faubert B, Villarino AV, O’Sullivan D, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–51.
Chapman NM, Boothby MR, Chi H. Metabolic coordination of T cell quiescence and activation. Nat Rev Immunol. 2020;20:55–70.
Franco F, Jaccard A, Romero P, Yu Y-R, Ho P-C. Metabolic and epigenetic regulation of T-cell exhaustion. Nat Metab. 2020;2:1001–12.
Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454.
Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–61.
Ma X, Bi E, Lu Y, Su P, Huang C, Liu L, et al. Cholesterol Induces CD8(+) T Cell Exhaustion in the Tumor Microenvironment. Cell Metab. 2019;30:143–56.e5.
Cao Y, Trillo-Tinoco J, Sierra RA, Anadon C, Dai W, Mohamed E, et al. ER stress-induced mediator C/EBP homologous protein thwarts effector T cell activity in tumors through T-bet repression. Nature Communications. 2019;10:1280.
Bengsch B, Johnson AL, Kurachi M, Odorizzi PM, Pauken KE, Attanasio J, et al. Bioenergetic Insufficiencies Due to Metabolic Alterations Regulated by the Inhibitory Receptor PD-1 Are an Early Driver of CD8(+) T Cell Exhaustion. Immunity. 2016;45:358–73.
Ogando J, Sáez ME, Santos J, Nuevo-Tapioles C, Gut M, Esteve-Codina A, et al. PD-1 signaling affects cristae morphology and leads to mitochondrial dysfunction in human CD8(+) T lymphocytes. J Immunother Cancer. 2019;7:151.
Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D, Long X, et al. Cancer cell-derived exosomal circUSP7 induces CD8(+) T cell dysfunction and anti-PD1 resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol Cancer. 2021;20:144.
Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y, et al. Exosome-derived circTRPS1 promotes malignant phenotype and CD8+ T cell exhaustion in bladder cancer microenvironments. Mol Ther. 2022;30:1054–70.
Mahat DB, Tippens ND, Martin-Rufino JD, Waterton SK, Fu J, Blatt SE, et al. Single-cell nascent RNA sequencing unveils coordinated global transcription. Nature. 2024;631:216–23.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583.
Goverde CA, Wolf B, Khakzad H, Rosset S, Correia BE. De novo protein design by inversion of the AlphaFold structure prediction network. Protein Sci. 2023;32:e4653.
Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630:493–500.
Wang J, Lisanza S, Juergens D, Tischer D, Watson JL, Castro KM, et al. Scaffolding protein functional sites using deep learning. Science. 2022;377:387–94.
Fudge JB. Diffusion model expands RoseTTAFold’s power. Nat Biotechnol. 2023;41:1072.
Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120.
Gao S, Liang X, Wang H, Bao B, Zhang K, Zhu Y, et al. Stem cell-like memory T cells: A perspective from the dark side. Cell Immunol. 2021;361:104273.
Lian J, Yue Y, Yu W, Zhang Y. Immunosenescence: a key player in cancer development. J Hematol Oncol. 2020;13:151.
Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015;12:1094–8.
Chen D, Lei EP. Function and regulation of chromatin insulators in dynamic genome organization. Curr Opin Cell Biol. 2019;58:61–8.
Wei GH, Liu DP, Liang CC. Chromatin ___domain boundaries: insulators and beyond. Cell Res. 2005;15:292–300.
Phillips-Cremins Jennifer E, Corces Victor G. Chromatin Insulators: Linking Genome Organization to Cellular Function. Mol Cell. 2013;50:461–74.
Author information
Authors and Affiliations
Contributions
Hao Zu, as the first author, proposed the research topic and conducted the literature search, analysis, illustration creation, and writing of the manuscript. Xiaoqin Chen, as the corresponding author, provided guidance throughout the writing process, integrated feedback, and reviewed and revised the final manuscript, as well as offering support with formatting revisions.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zu, H., Chen, X. Epigenetics behind CD8+ T cell activation and exhaustion. Genes Immun 25, 525–540 (2024). https://doi.org/10.1038/s41435-024-00307-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-024-00307-1