Abstract
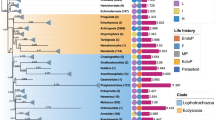
Studies of forces driving interlineage variability in the evolutionary rates (both sequence and architecture) of mitochondrial genomes often produce contradictory results. Flatworms (Platyhelminthes) exhibit the fastest-evolving mitogenomic sequences among all bilaterian phyla. To test the effects of multiple factors previously associated with different aspects of mitogenomic evolution, we used mitogenomes of 223 flatworm species, phylogenetic multilevel regression models, and causal inference. Thermic host environment (endothermic vs. ectothermic) had nonsignificant impacts on both sequence evolution and mitogenomic size. Mitogenomic gene order rearrangements (GORR) were mostly positively correlated with mitogenomic size (R2 ≈ 20–30%). Longevity was not (negatively) correlated with sequence evolution in flatworms. The predominantly free-living “turbellaria” exhibited much shorter branches and faster-evolving mitogenomic architecture than parasitic Neodermata. As a result, “parasitism” had a strong explanatory power on the branch length variability (>90%), and there was a negative correlation between GORR and branch length. However, the stem branch of Neodermata comprised 63.6% of the total average branch length. This evolutionary period was also marked by a high rate of gene order rearrangements in the ancestral Neodermata. We discuss how this period of rapid evolution deep in the evolutionary history may have decoupled sequence evolution rates from longevity and GORR, and overestimated the explanatory power of “parasitism”. This study shows that impacts of variables often vary across lineages, and stresses the importance accounting for the episodic nature of evolutionary patterns in studies of mitogenomic evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Allio R, Donega S, Galtier N, Nabholz B (2017) Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: Implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Mol Biol Evol 34:2762–2772
Arif S, Graham NAJ, Wilson S, MacNeil MA (2022) Causal drivers of climate-mediated coral reef regime shifts. Ecosphere 13:e3956
Bakke TA, Cable J, Harris PD (2007) The biology of Gyrodactylid Monogeneans: The “Russian-Doll Killers”. In: Baker J. R., Muller R., Rollinson D. (eds) Advances in Parasitology, Academic Press Vol 64, pp 161–460.
Bazin E, Glémin S, Galtier N (2006) Population size does not influence mitochondrial genetic diversity in animals. Science 312:570–572
Benesh DP, Parker G, Chubb JC (2021) Life-cycle complexity in helminths: What are the benefits? Evolution 75:1936–1952
Bernt M, Bleidorn C, Braband A, Dambach J, Donath A, Fritzsch G et al. (2013) A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol Phylogenet Evol 69:352–364
Bernt M, Merkle D, Ramsch K, Fritzsch G, Perseke M, Bernhard D et al. (2007) CREx: Inferring genomic rearrangements based on common intervals. Bioinformatics 23:2957–2958
Boore JL (2000) The Duplication/Random Loss Model for Gene Rearrangement Exemplified by Mitochondrial Genomes of Deuterostome Animals. In: Sankoff D., Nadeau J. H. (eds) Comparative Genomics: Empirical and Analytical Approaches to Gene Order Dynamics, Map Alignment and the Evolution of Gene Families, Springer Netherlands: Dordrecht, pp 133–147
Brabec J, Salomaki ED, Kolísko M, Scholz T, Kuchta R (2023) The evolution of endoparasitism and complex life cycles in parasitic Platyhelminths. Curr Biol 33:4269–4275.e3
Bürkner P-C (2018) Advanced Bayesian multilevel modeling with the R Package brms. R J 10:395–411
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973
Chong RA, Mueller RL (2013) Evolution along the mutation gradient in the dynamic mitochondrial genome of salamanders. Genome Biol Evol 5:1652–1660
Galtier N, Jobson RW, Nabholz B, Glémin S, Blier PU (2009) Mitochondrial whims: metabolic rate, longevity and the rate of molecular evolution. Biol Lett 5:413–416
Gillespie JH (1984) The molecular clock may be an episodic clock. Proc Natl Acad Sci 81:8009–8013
Giribet G, Distel DL, Polz M, Sterrer W, Wheeler WC (2000) Triploblastic relationships with emphasis on the acoelomates and the position of Gnathostomulida, Cycliophora, Plathelminthes, and Chaetognatha: A Combined Approach of 18S rDNA Sequences and Morphology. Syst Biol 49:539–562
Hahn C, Fromm B, Bachmann L (2014) Comparative genomics of Flatworms (Platyhelminthes) reveals shared genomic features of ecto- and endoparastic Neodermata. Genome Biol Evol 6:1105–1117
Hassanin A (2006) Phylogeny of Arthropoda inferred from mitochondrial sequences: Strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol Phylogenet Evol 38:100–116
Hu F, Lin Y, Tang J (2014) MLGO: phylogeny reconstruction and ancestral inference from gene-order data. BMC Bioinforma 15:354
Hua X, Cowman P, Warren D, Bromham L (2015) Longevity is linked to mitochondrial mutation rates in Rockfish: A test using Poisson Regression. Mol Biol Evol 32:2633–2645
Hyman BC, Lewis SC, Tang S, Wu Z (2011) Rampant gene rearrangement and haplotype hypervariation among nematode mitochondrial genomes. Genetica 139:611–615
Jackson JA, Tinsley RC (1998) Effects of temperature on oviposition rate in Protopolystoma xenopodis (Monogenea: Polystomatidae). Int J Parasitol 28:309–315
Jakovlić I, Zou H, Chen J-H, Lei H-P, Wang G-T, Liu J et al. (2021) Slow crabs - fast genomes: locomotory capacity predicts skew magnitude in crustacean mitogenomes. Mol Ecol 30:5488–5502
Jakovlić I, Zou H, Ye T, Zhang H, Liu X, Xiang C-Y et al. (2023) Mitogenomic evolutionary rates in Bilateria are influenced by parasitic lifestyle and locomotory capacity. Nat Commun 14:6307
Jeschke JM, Kokko H (2009) The roles of body size and phylogeny in fast and slow life histories. Evol Ecol 23:867–878
Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30:772–780
Kosakovsky Pond SL, Poon AFY, Velazquez R, Weaver S, Hepler NL, Murrell B et al. (2020) HyPhy 2.5—A customizable platform for evolutionary hypothesis testing using Phylogenies. Mol Biol Evol 37:295–299
Lagisz M, Poulin R, Nakagawa S (2013) You are where you live: parasitic nematode mitochondrial genome size is associated with the thermal environment generated by hosts. J Evol Biol 26:683–690
Lajbner Z, Pnini R, Camus MF, Miller J, Dowling DK (2018) Experimental evidence that thermal selection shapes mitochondrial genome evolution. Sci Rep. 8:9500
Lanfear R, Thomas JA, Welch JJ, Brey T, Bromham L (2007) Metabolic rate does not calibrate the molecular clock. Proc Natl Acad Sci 104:15388–15393
Laumer CE, Fernández R, Lemer S, Combosch D, Kocot KM, Riesgo A et al. (2019) Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc R Soc B Biol Sci 286:20190831
Laumer CE, Giribet G (2014) Inclusive taxon sampling suggests a single, stepwise origin of ectolecithality in Platyhelminthes. Biol J Linn Soc 111:570–588
Letunic I, Bork P (2024) Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res 52:W78–W82
Li W-H, Tanimura M, Sharp PM (1987) An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol 25:330–342
Lunt DH, Hyman BC (1997) Animal mitochondrial DNA recombination. Nature 387:247–247
Lynch M, Koskella B, Schaack S (2006) Mutation pressure and the evolution of organelle genomic architecture. Science 311:1727–1730
Martin AP, Palumbi SR (1993) Body size, metabolic rate, generation time, and the molecular clock. Proc Natl Acad Sci 90:4087–4091
Min XJ, Hickey DA (2008) An evolutionary footprint of age-related natural selection in mitochondrial DNA. J Mol Evol 67:412
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A et al. (2020) IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534
Muller R, Wakelin D (2002) Worms and human disease, 2nd edn. CABi Publishing: Wallingford, UK.
Nabholz B, Glémin S, Galtier N (2008a) Strong variations of mitochondrial mutation rate across mammals—the longevity hypothesis. Mol Biol Evol 25:120–130
Nabholz B, Glémin S, Galtier N (2009) The erratic mitochondrial clock: variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals. BMC Evol Biol 9:54
Nabholz B, Lanfear R, Fuchs J (2016) Body mass-corrected molecular rate for bird mitochondrial DNA. Mol Ecol 25:4438–4449
Nabholz B, Mauffrey J-F, Bazin E, Galtier N, Glemin S (2008b) Determination of mitochondrial genetic diversity in mammals. Genetics 178:351–361
Perkins E (2010) Family ties: molecular phylogenetics, evolution and radiation of flatworm parasites (Monogenea: capsalidae). PhD Thesis, University of Adelaide, School of Earth and Environmental Sciences
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41:353–358
Pinheiro J, Bates D (2006) Mixed-effects models in S and S-PLUS, 1st edn. Springer Science & Business Media: New York, NY.
Pinheiro J, Bates D, R Core Team (2024) nlme: Linear and Nonlinear Mixed Effects Models (https://CRAN.R-project.org/package=nlme)
Podsiadlowski L, Braband A, Struck TH, von Döhren J, Bartolomaeus T (2009) Phylogeny and mitochondrial gene order variation in Lophotrochozoa in the light of new mitogenomic data from Nemertea. BMC Genomics 10:364
Poulin R, Latham ADM (2003) Effects of initial (larval) size and host body temperature on growth in trematodes. Can J Zool 81:574–581
Poulin R, Morand S (2000) The diversity of parasites. Q Rev Biol 75:277–293
Rand DM (1993) Endotherms, ectotherms, and mitochondrial genome-size variation. J Mol Evol 37:281–295
Rand DM (1994) Thermal habit, metabolic rate and the evolution of mitochondrial DNA. Trends Ecol Evol 9:125–131
Rohde K (1994) The origins of parasitism in the Platyhelminthes. Int J Parasitol 24:1099–1115
Rosa MT, Oliveira DS, Loreto ELS (2017) Characterization of the first mitochondrial genome of a catenulid flatworm: Stenostomum leucops (Platyhelminthes). J Zool Syst Evol Res 55:98–105
Saclier N, François CM, Konecny-Dupré L, Lartillot N, Guéguen L, Duret L et al. (2018) Life history traits impact the nuclear rate of substitution but not the mitochondrial rate in isopods. Mol Biol Evol 35:2900–2912
Shao R, Dowton M, Murrell A, Barker SC (2003) Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol Biol Evol 20:1612–1619
Shtolz N, Mishmar D (2023) The metazoan landscape of mitochondrial DNA gene order and content is shaped by selection and affects mitochondrial transcription. Commun Biol 6:1–15
Solà E, Álvarez-Presas M, Frías-López C, Littlewood DTJ, Rozas J, Riutort M (2015) Evolutionary analysis of mitogenomes from parasitic and free-living flatworms. PLOS ONE 10:e0120081
Struck TH, Golombek A, Hoesel C, Dimitrov D, Elgetany AH (2023) Mitochondrial genome evolution in Annelida—A systematic study on conservative and variable gene orders and the factors influencing its evolution. Syst Biol 72:925–945
Struck TH, Wey-Fabrizius AR, Golombek A, Hering L, Weigert A, Bleidorn C et al. (2014) Platyzoan paraphyly based on phylogenomic data supports a noncoelomate ancestry of Spiralia. Mol Biol Evol 31:1833–1849
Tan MH, Gan HM, Lee YP, Bracken-Grissom H, Chan T-Y, Miller AD et al. (2019) Comparative mitogenomics of the Decapoda reveals evolutionary heterogeneity in architecture and composition. Sci Rep. 9:1–16
Textor J, van der Zander B, Gilthorpe MS, Liśkiewicz M, Ellison GT (2016) Robust causal inference using directed acyclic graphs: the R package ‘dagitty’. Int J Epidemiol 45:1887–1894
Therneau T (2018) The lmekin function (https://cran.r-project.org/web/packages/coxme/vignettes/lmekin.pdf) (Accessed 17.7.2024.)
Thomas JA, Welch JJ, Lanfear R, Bromham L (2010) A generation time effect on the rate of molecular evolution in invertebrates. Mol Biol Evol 27:1173–1180
Thomas JA, Welch JJ, Woolfit M, Bromham L (2006) There is no universal molecular clock for invertebrates, but rate variation does not scale with body size. Proc Natl Acad Sci 103:7366–7371
Valenzano DR, Aboobaker A, Seluanov A, Gorbunova V (2017) Non-canonical aging model systems and why we need them. EMBO J 36:959–963
Welch JJ, Bininda-Emonds OR, Bromham L (2008) Correlates of substitution rate variation in mammalian protein-coding sequences. BMC Evol Biol 8:53
Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K (2015) RELAX: Detecting relaxed selection in a phylogenetic framework. Mol Biol Evol 32:820–832
Xiang C, Gao F, Jakovlić I, Lei H, Hu Y, Zhang H et al. (2023) Using PhyloSuite for molecular phylogeny and tree‐based analyses. iMeta 2:e87
Xu W, Jameson D, Tang B, Higgs PG (2006) The relationship between the rate of molecular evolution and the rate of genome rearrangement in animal mitochondrial genomes. J Mol Evol 63:375–392
Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX et al. (2020) PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour 20:348–355
Zhang D, Jakovlić I, Zou H, Liu F, Xiang C-Y, Gusang Q et al. (2024) Strong mitonuclear discordance in the phylogeny of Neodermata and evolutionary rates of Polyopisthocotylea. Int J Parasitol 54:213–223
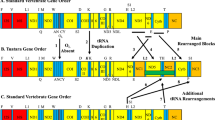
Zhang D, Li WX, Zou H, Wu SG, Li M, Jakovlić I et al. (2019) Homoplasy or plesiomorphy? Reconstruction of the evolutionary history of mitochondrial gene order rearrangements in the subphylum Neodermata. Int J Parasitol 49:819–829
Zou H, Chen F-L, Li W-X, Li M, Lei H-P, Zhang D et al. (2022a) Inverted base composition skews and discontinuous mitochondrial genome architecture evolution in the Enoplea (Nematoda). BMC Genomics 23:376
Zou H, Lei H-P, Chen R, Chen F-L, Li W-X, Li M et al. (2022b) Evolutionary rates of mitochondrial sequences and gene orders in Spirurina (Nematoda) are episodic but synchronised. Water Biol Secur 1:100033
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32360927, 32102840); the Key Project of Natural Science Foundation of Tibet (XZ202301ZR0028G); the Open Fund Project of Key Laboratory of Breeding Biotechnology and Sustainable Aquaculture (2023FB07); and the Start-up Funds of Introduced Talent in Lanzhou University (561120206). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We would like to thank Dr. Xiang Liu for helpful discussions and technical assistance.
Author information
Authors and Affiliations
Contributions
IJ: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing – original draft. TY, FZ, YS, YM: formal analysis, investigation, methodology, writing – review & editing. HZ: formal analysis, resources, validation, visualization, writing – review & editing. GTW: formal analysis, resources, validation, visualization, supervision, writing – review & editing. WXL: validation, funding acquisition, supervision, writing – review & editing. ZD: data curation, formal analysis, investigation, methodology, resources, software, validation, visualization, writing – review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Research ethics statement
No approval of research ethics committees was required to accomplish the goals of this study because it was conducted on previously sequenced mitogenomes acquired from GenBank and included only unregulated invertebrate species.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Xiangjiang Zhan.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jakovlić, I., Ye, T., Zou, H. et al. Drivers of interlineage variability in mitogenomic evolutionary rates in Platyhelminthes. Heredity 133, 276–286 (2024). https://doi.org/10.1038/s41437-024-00712-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-024-00712-2