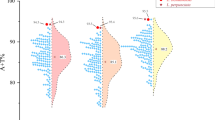
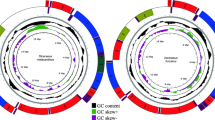
Abstract
Feliform carnivores face dual threats from habitat fragmentation and climate change, but unresolved phylogenetic relationships and unclear adaptive mechanisms hinder the development of conservation strategies. This study integrates mitochondrial genome data from 75 extant species (including three newly obtained taxa: Helogale parvula, Suricata suricatta, and Neofelis diardi) to resolve taxonomic controversies and reveal adaptive evolutionary mechanisms. Bayesian phylogenetic reconstruction strongly supports a sister-group relationship between Felidae and Prionodontidae (posterior probability PP = 1.0), overturning traditional morphological classifications. Divergence time estimation indicates that the crown group of Feliformia originated in the Middle Eocene (46 Ma), with key radiation events synchronized with Oligocene-Miocene climatic upheavals and continental collisions. Adaptive evolution analyses show that mitochondrial protein-coding genes (PCGs) are predominantly under purifying selection. However, significant positive selection signals were detected in the ND4 gene of Nandinia binotata and the COX2 gene of Pantherinae, potentially linked to arid adaptation and predatory energy demands, respectively. The frequent use of GTG start codons in the COX1 gene of Neofelis diardi suggests metabolic fine-tuning for island ecosystems. Conservation genomics identifies Prionodon pardicolor and Neofelis nebulosa as Evolutionarily Significant Units (ESUs) with heightened vulnerability to habitat fragmentation. By integrating mitogenomic architecture, deep-time biogeography, and contemporary selection pressures, this study establishes a unified framework bridging molecular systematics and conservation strategies, providing scientific guidance for protecting rapidly evolving lineages.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Alexanian A, Sorokin A (2017) Cyclooxygenase 2: protein-protein interactions and posttranslational modifications. Physiol Genom 49(11):667–681.
Awadi A, Ben Slimen H, Schaschl H, Knauer F, Suchentrunk F (2021) Positive selection on two mitochondrial coding genes and adaptation signals in hares (genus Lepus) from China. BMC Ecol Evol 21(1):100.
Barnett R, Barnes I, Phillips MJ, Martin LD, Harington CR, Leonard JA et al. (2005) Evolution of the extinct Sabretooths and the American cheetah-like cat. Curr Biol 15(15):R589–R590.
Barycka E (2007) Evolution and systematics of the feliform Carnivora. Mamm Biol 72(5):257–282.
Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G et al. (2013) MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol 69(2):313–319.
Bininda-Emonds OR, Gittleman JL, Purvis A (1999) Building large trees by combining phylogenetic information: a complete phylogeny of the extant Carnivora (Mammalia). Biol Rev 74(2):143–175.
Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27(8):1767–1780.
Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A et al. (2019) BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol 15(4):e1006650.
Broggini C, Cavallini M, Vanetti I, Abell J, Binelli G, Lombardo G (2024) From Caves to the Savannah, the Mitogenome History of Modern Lions (Panthera leo) and Their Ancestors. Int J Mol Sci 25(10):5193.
Burgin CJ, Colella JP, Kahn PL, Upham NS (2018) How many species of mammals are there?. J Mammal 99(1):1–14.
Bursell MG, Dikow RB, Figueiró HV, Dudchenko O, Flanagan JP, Aiden EL et al. (2022) Whole genome analysis of clouded leopard species reveals an ancient divergence and distinct demographic histories. iScience 25(12):105647.
Clyde D (2019) Decoding dragon DNA. Nat Rev Genet 20(10):564–565.
Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res 45(4):e18.
Estes JA, Terborgh J, Brashares JS, Power ME, Berger J, Bond WJ et al. (2011) Trophic downgrading of planet Earth. Science 333(6040):301–306.
Feng S, Bai M, Rivas-González I, Li C, Liu S, Tong Y et al. (2022) Incomplete lineage sorting and phenotypic evolution in marsupials. Cell 185(10):1646–1660.e1618.
Figueiró HV, Li G, Trindade FJ, Assis J, Pais F, Fernandes G et al. (2017) Genome-wide signatures of complex introgression and adaptive evolution in the big cats. Sci Adv 3(7):e1700299.
Galtier N, Nabholz B, Glémin S, Hurst GD (2009) Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol 18(22):4541–4550.
Hassanin A, Veron G, Ropiquet A, Jansen van Vuuren B, Lécu A, Goodman SM et al. (2021) Evolutionary history of Carnivora (Mammalia, Laurasiatheria) inferred from mitochondrial genomes. PLoS ONE 16(2):e0240770.
Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A, Teeling E et al. (2006) The late Miocene radiation of modern Felidae: a genetic assessment. Science 311(5757):73–77.
Kitchener A, Breitenmoser C, Eizirik E, Gentry A, Werdelin L, Wilting A et al. (2017) A revised taxonomy of the Felidae. The final report of the Cat Classification Task Force of the IUCN/SSC Cat Specialist Group. Cat News Special Issue: 80 pp.
Koufos GD (2022) The Fossil Record of Hyaenids (Mammalia: Carnivora: Hyaenidae) in Greece. In: Vlachos E (ed) Fossil Vertebrates of Greece Vol. 2: Laurasiatherians, Artiodactyles, Perissodactyles, Carnivorans, and Island Endemics. Springer International Publishing: Cham, pp 555-576.
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2017) PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol Biol Evol 34(3):772–773.
Miralles A, Puillandre N, Vences M (2024) DNA Barcoding in Species Delimitation: From Genetic Distances to Integrative Taxonomy. Methods Mol Biol 2744:77–104.
Nyakatura K, Bininda-Emonds ORP (2012) Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biol 10(1):12.
Parvathy ST, Udayasuriyan V, Bhadana V (2022) Codon usage bias. Mol Biol Rep 49(1):539–565.
Polly PD (2001) Paleontology and the comparative method: ancestral node reconstructions versus observed node values. Am Nat 157(6):596–609.
Popadin KY, Nikolaev SI, Junier T, Baranova M, Antonarakis SE (2013) Purifying selection in mammalian mitochondrial protein-coding genes is highly effective and congruent with evolution of nuclear genes. Mol Biol Evol 30(2):347–355.
de Queiroz A (2005) The resurrection of oceanic dispersal in historical biogeography. Trends Ecol Evol 20(2):68–73.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S et al. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542.
Rozewicki J, Li S, Amada KM, Standley DM, Katoh K (2019) MAFFT-DASH: integrated protein sequence and structural alignment. Nucl Acids Res 47(W1):W5–W10.
Rundell RJ, Price TD (2009) Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends Ecol Evol 24(7):394–399.
Seehausen O (2015) Process and pattern in cichlid radiations - inferences for understanding unusually high rates of evolutionary diversification. N Phytol 207(2):304–312.
Sen S (2013) Dispersal of African mammals in Eurasia during the Cenozoic: Ways and whys. Geobios 46(1):159–172.
Shackleton S, Seltzer A, Baggenstos D, Lisiecki LE (2023) Benthic δ18O records Earth’s energy imbalance. Nat Geosci 16(9):797–802.
Solaini G, Baracca A, Lenaz G, Sgarbi G (2010) Hypoxia and mitochondrial oxidative metabolism. Biochimica et Biophysica Acta (BBA) - Bioenerg 1797(6):1171–1177.
Strömberg CAE (2011) Evolution of grasses and grassland ecosystems. Annu Rev Earth Planet Sci 39(39):517–544.
Summerer M, Horst J, Erhart G, Weißensteiner H, Schönherr S, Pacher D et al. (2014) Large-scale mitochondrial DNA analysis in Southeast Asia reveals evolutionary effects of cultural isolation in the multi-ethnic population of Myanmar. BMC Evol Biol 14(1):17.
Sunnucks P, Morales HE, Lamb AM, Pavlova A, Greening C (2017) Integrative approaches for studying mitochondrial and nuclear genome co-evolution in oxidative phosphorylation. Front Genet 8:25.
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7):3022–3027.
Wang B, Jia P, Gao S, Zhao H, Zheng G, Xu L et al. (2025) Long and accurate: how HiFi sequencing is transforming genomics. Genom Proteom Bioinf 7:qzaf003.
Wang Q, Wu Y, Merchant A, Li E, Wei M, Zhang Y et al. (2024) The mitochondrial genome and life history of Tomostethus sinofraxini (Hymenoptera: Tenthredinidae), an emerging pest of Fraxinus chinensis. J Econ Entomol 117(2):564–577.
Wang X, Shang Y, Wu X, Wei Q, Zhou S, Sun G et al. (2023) Divergent evolution of mitogenomics in Cetartiodactyla niche adaptation. Org Divers Evol 23(1):243–259.
Wang X, Zhou S, Wu X, Wei Q, Shang Y, Sun G et al. (2021) High-altitude adaptation in vertebrates as revealed by mitochondrial genome analyses. Ecol Evol 11(21):15077–15084.
Wei L, He J, Jia X, Qi Q, Liang Z, Zheng H et al. (2014) Analysis of codon usage bias of mitochondrial genome in Bombyx moriand its relation to evolution. BMC Evol Biol 14(1):262.
Yang L, Wei F, Zhan X, Fan H, Zhao P, Huang G et al. (2022) Evolutionary conservation genomics reveals recent speciation and local adaptation in threatened takins. Mol Biol Evol 39(6):msac111.
Yang Z (2007) PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol 24(8):1586–1591.
Zhang WQ, Zhang MH (2013) Complete mitochondrial genomes reveal phylogeny relationship and evolutionary history of the family Felidae. Genet Mol Res : GMR 12(3):3256–3262.
Zhou Y, Wang S-R, Ma J-Z (2017) Comprehensive species set revealing the phylogeny and biogeography of Feliformia (Mammalia, Carnivora) based on mitochondrial DNA. PLoS ONE 12(3):e0174902.
Zwonitzer KD, Iverson ENK, Sterling JE, Weaver RJ, Maclaine BA, Havird JC (2023) Disentangling positive selection from relaxed selection in animal mitochondrial genomes. Am Nat 202(4):E121–e129.
Funding
This research was funded by the National Natural Science Foundation of China (32470448, 32270444), the Youth Innovation Team of Shandong Universities (2022KJ177).
Author information
Authors and Affiliations
Contributions
Xiaoyang Wu, Yamin Xing and Xibao Wang conducted the project, collected the fitness and copy-number data, interpreted results, and wrote the initial draft of the manuscript. Honghai Zhang generated the original experimental line, provided funding, made several figures and helped write the later versions of the manuscript. Yongquan Shang, Yao Chen, Mingke Han and Weilai Sha provided funding, designed the experiments and helped write the later versions of the manuscript. All authors contributed to the final revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Research ethics statement
The culturing is not associated with any research ethics constraints.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Xiangjiang Zhan.
Supplementary information
41437_2025_772_MOESM3_ESM.xlsx
Table S3: Test for positive selection in divergent clades of suborder Feliformia mtPCGs with branch model (*P < 0.05; **P < 0.01)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, X., Xing, Y., Wang, X. et al. Mitogenomic resolution of phylogenetic conflicts and adaptive signatures in feliform carnivorans. Heredity (2025). https://doi.org/10.1038/s41437-025-00772-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41437-025-00772-y