Abstract
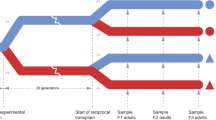
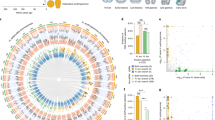
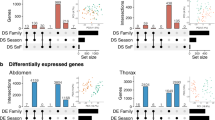
Inducible defences in response to predation risk are a well-known example of adaptive phenotypic plasticity. Although inducible defences have been studied mainly within a generation (within-generational plasticity), there is now clear evidence that ancestral exposure to predation risk can influence the defences expressed by offspring, even if they have not been exposed themselves (transgenerational plasticity). The molecular mechanisms allowing the transmission of environmental information across generations are not well understood. In this study, we combined measures of antipredator responses (behavioural and morphological) with transcriptomic investigations across two generations in the freshwater snail Physa acuta. We hypothesised that both within- and transgenerational plasticity would induce phenotypic changes associated with differential gene expression. Our results confirmed within- and transgenerational plasticity: F1 snails respond to predator-cue exposure by increasing escape behaviour, reducing shell length, and developing thicker and slenderer shells, whereas F2 snails from exposed parents have longer and thicker shells with narrower apertures. Within- and transgenerational plasticity were accompanied by the differential expression of 112 genes (101 up- and 11 downregulated) and 23 differentially expressed genes (17 up- and 6 downregulated), respectively. Within- and transgenerational plasticity did not share common differentially expressed genes, but the associated molecular functions, involving metabolism and transcription regulation, were similar. These results suggest that predator-induced within-generational plasticity and transgenerational plasticity may result from different genomic pathways and may evolve independently.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA-seq data have been deposited in the European Nucleotide Archive (ENA) under the accession number [PRJEB87216]. The raw sequencing data are publicly available at. Additionally, scripts used for the analysis, along with the non-genomic data, have been made available on Zenodo. The repository can be accessed at https://doi.org/10.5281/zenodo.15043538.
References
Agrawal AA, Laforsch C, Tollrian R (1999) Transgenerational induction of defences in animals and plants. Nature 401(6748):6748. https://doi.org/10.1038/43425
Aubin-Horth N, Renn SCP (2009) Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol Ecol 18(18):3763–3780. https://doi.org/10.1111/j.1365-294X.2009.04313.x
Auld JR, Houser R (2015) Age-dependent effects of predation risk on reproductive success in a freshwater snail. Evolution 69(10):2793–2798. https://doi.org/10.1111/evo.12769
Auld JR, Relyea RA (2011) Adaptive plasticity in predator-induced defenses in a common freshwater snail: altered selection and mode of predation due to prey phenotype. Evol Ecol 25(1):189–202. https://doi.org/10.1007/s10682-010-9394-1
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting Linear Mixed-Effects Models Using lme4. J Stat Softw 67(1):1–48. https://doi.org/10.18637/jss.v067.i01
Beaty LE, Wormington JD, Kensinger BJ, Bayley KN, Goeppner SR, Gustafson KD, Luttbeg B (2016) Shaped by the past, acting in the present: transgenerational plasticity of anti-predatory traits. Oikos 125(11):1570–1576. https://doi.org/10.1111/oik.03114
Bell AM, Hellmann JK (2019) An integrative framework for understanding the mechanisms and multigenerational consequences of transgenerational plasticity. Annu Rev Ecol Evol Syst 50(1):97–118. https://doi.org/10.1146/annurev-ecolsys-110218-024613
Bell AM, Stein LR (2017) Transgenerational and developmental plasticity at the molecular level: lessons from Daphnia. Mol Ecol 26(19):4859–4861. https://doi.org/10.1111/mec.14327
Blackwell TK, Sewell AK, Wu Z, Han M (2019) TOR signaling in caenorhabditis elegans development, metabolism, and aging. Genetics 213(2):329–360. https://doi.org/10.1534/genetics.119.302504
Blum M, Andreeva A, Florentino LC, Chuguransky SR, Grego T, Hobbs E, Pinto BL, Orr A, Paysan-Lafosse T, Ponamareva I, Salazar GA, Bordin N, Bork P, Bridge A, Colwell L, Gough J, Haft DH, Letunic I, Llinares-López F, Bateman A (2024) InterPro: The protein sequence classification resource in 2025. Nucleic Acids Res 53(D1):D444–D456. https://doi.org/10.1093/nar/gkae1082
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34(5):Article 5. https://doi.org/10.1038/nbt.3519
Bukowski SJ, Auld JR (2014) The effects of calcium in mediating the inducible morphological defenses of a freshwater snail, Physa acuta. Aquat Ecol 48(1):85–90. https://doi.org/10.1007/s10452-013-9468-6
Burton T, Metcalfe NB (2014) Can environmental conditions experienced in early life influence future generations?. Proc R Soc B Biol Sci 281(1785):20140311. https://doi.org/10.1098/rspb.2014.0311
Clark MS, Suckling CC, Cavallo A, Mackenzie CL, Thorne MAS, Davies AJ, Peck LS (2019) Molecular mechanisms underpinning transgenerational plasticity in the green sea urchin Psammechinus miliaris. Sci Rep https://www-nature-com.docelec.univ-lyon1.fr/articles/s41598-018-37255-6
Colicchio JM, Herman J (2020) Empirical patterns of environmental variation favor adaptive transgenerational plasticity. Ecol Evol 10(3):1648–1665. https://doi.org/10.1002/ece3.6022
Danchin É, Charmantier A, Champagne FA, Mesoudi A, Pujol B, Blanchet S (2011) Beyond DNA: integrating inclusive inheritance into an extended theory of evolution. Nat Rev Genet 12(7):Article 7. https://doi.org/10.1038/nrg3028
DeWitt TJ (1998) Costs and limits of phenotypic plasticity: tests with predator-induced morphology and life history in a freshwater snail. J Evol Biol 11(4):465–480. https://doi.org/10.1046/j.1420-9101.1998.11040465.x
Duncan EJ, Gluckman PD, Dearden PK (2014) Epigenetics, plasticity, and evolution: how do we link epigenetic change to phenotype?. J Exp Zool Part B Mol Dev Evol 322(4):208–220. https://doi.org/10.1002/jez.b.22571
Ezard THG, Prizak R, Hoyle RB (2014) The fitness costs of adaptation via phenotypic plasticity and maternal effects. Funct Ecol 28(3):693–701
Fallet M, Luquet E, David P, Cosseau C (2020) Epigenetic inheritance and intergenerational effects in mollusks. Gene 729:144166. https://doi.org/10.1016/j.gene.2019.144166
Galloway LF, Etterson JR (2007) Transgenerational plasticity is adaptive in the wild. Science 318(5853):1134–1136. https://doi.org/10.1126/science.1148766
Gilbert SF (2001) Ecological developmental biology: developmental biology meets the real world. Dev Biol 233(1):1–12. https://doi.org/10.1006/dbio.2001.0210
Gustafson K, Kensinger B, Bolek M, Luttbeg B (2014) Distinct snail (Physa) morphotypes from different habitats converge in shell shape and size under common garden conditions. Evolut Ecol Res 16:77–89
Hales NR, Schield DR, Andrew AL, Card DC, Walsh MR, Castoe TA (2017) Contrasting gene expression programs correspond with predator-induced phenotypic plasticity within and across generations in Daphnia. Mol Ecol 26(19):5003–5015. https://doi.org/10.1111/mec.14213
Hammer O, Harper DAT, Ryan PD (2001) PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron
Herman J, Sultan S (2011) Adaptive transgenerational plasticity in plants: case studies, mechanisms, and implications for natural populations. Front Plant Sci 2. https://www.frontiersin.org/articles/10.3389/fpls.2011.00102
Jablonka E, Raz G (2009) Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol 84(2):131–176. https://doi.org/10.1086/598822
Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. https://doi.org/10.1093/bioinformatics/btu031
Katewa SD, Kapahi P (2011) Role of TOR signaling in aging and related biological processes in Drosophila melanogaster. Exp Gerontol 46(5):382. https://doi.org/10.1016/j.exger.2010.11.036
Khorchid A, Ikura M (2002) How calpain is activated by calcium. Nat Struct Biol 9(4):239–241. https://doi.org/10.1038/nsb0402-239
Kikuchi DW, Allen WL, Arbuckle K, Aubier TG, Briolat ES, Burdfield-Steel ER, Cheney KL, Daňková K, Elias M, Hämäläinen L, Herberstein ME, Hossie TJ, Joron M, Kunte K, Leavell BC, Lindstedt C, Lorioux-Chevalier U, McClure M, McLellan CF, Exnerová A (2023) The evolution and ecology of multiple antipredator defences. J Evol Biol 36(7):975–991. https://doi.org/10.1111/jeb.14192
Kuijper B, Hoyle RB (2015) When to rely on maternal effects and when on phenotypic plasticity? Evolution 69(4):950–968. https://doi.org/10.1111/evo.12635
Kuznetsova A, Brockhoff PB, Christensen RHB (2017) lmertest package: tests in linear mixed effects models. J Stat Software 82(13). https://doi.org/10.18637/jss.v082.i13
Ledón-Rettig CC (2023) A transcriptomic investigation of heat-induced transgenerational plasticity in beetles. Biol J Linn Soc 138(3):318–327. https://doi.org/10.1093/biolinnean/blac151
Lee PC, Bussière LF, Webber CE, Poole JH, Moss CJ (2013) Enduring consequences of early experiences: 40 year effects on survival and success among African elephants (Loxodonta Africana). Biol Lett 9(2):20130011. https://doi.org/10.1098/rsbl.2013.0011
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Luquet E, Léna J-P, David P, Joly P, Lengagne T, Perrin N, Plénet S (2011) Consequences of genetic erosion on fitness and phenotypic plasticity in European tree frog populations (Hyla arborea). J Evol Biol 24(1):99–110. https://doi.org/10.1111/j.1420-9101.2010.02138.x
Luquet E, Tariel J (2016) Offspring reaction norms shaped by parental environment: Interaction between within- and trans-generational plasticity of inducible defenses. BMC Evol Biol 16(1):1. https://doi.org/10.1186/s12862-016-0795-9
MacLeod KJ, Monestier C, Ferrari MCO, McGhee KE, Sheriff MJ, Bell AM (2022) Predator-induced transgenerational plasticity in animals: a meta-analysis. Oecologia 200(3):371–383. https://doi.org/10.1007/s00442-022-05274-w
Mommer BC, Bell AM (2014) Maternal experience with predation risk influences genome-wide embryonic gene expression in threespined sticklebacks (Gasterosteus aculeatus). PloS One 9(6):e98564. https://doi.org/10.1371/journal.pone.0098564
Orsini L, Brown JB, Shams Solari O, Li D, He S, Podicheti R, Stoiber MH, Spanier KI, Gilbert D, Jansen M, Rusch DB, Pfrender ME, Colbourne JK, Frilander MJ, Kvist J, Decaestecker E, De Schamphelaere KAC, De Meester L (2018) Early transcriptional response pathways in Daphnia magna are coordinated in networks of crustacean-specific genes. Mol Ecol 27(4):886–897. https://doi.org/10.1111/mec.14261
Pigliucci M (2005) Evolution of phenotypic plasticity: where are we going now? Trends Ecol Evol 20(9):481–486. https://doi.org/10.1016/j.tree.2005.06.001
Prizak R, Ezard THG, Hoyle RB (2014) Fitness consequences of maternal and grandmaternal effects. Ecol Evol 4(15):3139–3145. https://doi.org/10.1002/ece3.1150
Reger J, Lind MI, Robinson MR, Beckerman AP (2018) Predation drives local adaptation of phenotypic plasticity. Nat Ecol Evol 2(1):100–107. https://doi.org/10.1038/s41559-017-0373-6
Rozenberg A, Parida M, Leese F, Weiss LC, Tollrian R, Manak JR (2015) Transcriptional profiling of predator-induced phenotypic plasticity in Daphnia pulex. Front Zool 12(1):18. https://doi.org/10.1186/s12983-015-0109-x
R Studio Team (2020) RStudio: Integrated Development Environment for R
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7):671–675
Segers FHID, Taborsky B (2011) Juvenile exposure to predator cues induces a larger egg size in fish. Proc R Soc B Biol Sci 279(1731):1241–1248. https://doi.org/10.1098/rspb.2011.1290
Sharda S, Zuest T, Erb M, Taborsky B (2021) Predator-induced maternal effects determine adaptive antipredator behaviors via egg composition. Proc Natl Acad Sci 118(37):e2017063118. https://doi.org/10.1073/pnas.2017063118
Skottene E, Tarrant AM, Altin D, Olsen RE, Choquet M, Kvile KØ (2020) Lipid metabolism in Calanus finmarchicus is sensitive to variations in predation risk and food availability. Sci Rep 10(1):22322. https://doi.org/10.1038/s41598-020-79165-6
Slos S, Stoks R (2008) Predation risk induces stress proteins and reduces antioxidant defense. Funct Ecol 22(4):637–642. https://doi.org/10.1111/j.1365-2435.2008.01424.x
Smithson M, Thorson JLM, Sadler-Riggleman I, Beck D, Skinner MK, Dybdahl M (2020) Between-generation phenotypic and epigenetic stability in a clonal snail. Genome Biol Evol 12(9):1604–1615. https://doi.org/10.1093/gbe/evaa181
Stein LR, Bukhari SA, Bell AM (2018) Personal and transgenerational cues are nonadditive at the phenotypic and molecular level. Nat Ecol Evol 2(8):1306–1311. https://doi.org/10.1038/s41559-018-0605-4
Tariel J, Luquet É, Plénet S (2020) Interactions between maternal, paternal, developmental, and immediate environmental effects on anti-predator behavior of the Snail Physa acuta. Front Ecol Evol 8. https://www.frontiersin.org/articles/10.3389/fevo.2020.591074
Tariel J, Plénet S, Luquet E (2020a) How do developmental and parental exposures to predation affect personality and immediate behavioural plasticity in the snail Physa acuta? Proc R Soc B Biol Sci 287(1941):20201761. https://doi.org/10.1098/rspb.2020.1761
Tariel J, Plénet S, Luquet É (2020b) Transgenerational plasticity in the context of predator-prey interactions. Front Ecol Evol 8. https://doi.org/10.3389/fevo.2020.548660
Tariel J, Plénet S, Luquet É (2020c) Transgenerational plasticity of inducible defences: combined effects of grand-parental, parental and current environments. Ecol Evol 10(5):2367–2376. https://doi.org/10.1002/ece3.6046
Tariel-Adam J, Luquet É, Plénet S (2023) Sensitive windows for within- and trans-generational plasticity of anti-predator defences. Peer Community J 3. https://doi.org/10.24072/pcjournal.304
Thorson JLM, Smithson M, Beck D, Sadler-Riggleman I, Nilsson E, Dybdahl M, Skinner MK (2017) Epigenetics and adaptive phenotypic variation between habitats in an asexual snail. Sci Rep 7(1):14139. https://doi.org/10.1038/s41598-017-14673-6
Thorson JLM, Smithson M, Sadler-Riggleman I, Beck D, Dybdahl M, Skinner MK (2019) Regional epigenetic variation in asexual snail populations among urban and rural lakes. Environ Epigenet 5(4):dvz020. https://doi.org/10.1093/eep/dvz020
Tollrian R (1995) Predator-induced morphological defenses: costs, life history shifts, and maternal effects in Daphnia pulex. Ecology 76(6):1691–1705. https://doi.org/10.2307/1940703
Turner AM, Fetterolf SA, Bernot RJ (1999) Predator identity and consumer behavior: differential effects of fish and crayfish on the habitat use of a freshwater snail. Oecologia 118(2):242–247. https://doi.org/10.1007/s004420050724
Viney M, Diaz A (2012) Phenotypic plasticity in nematodes: evolutionary and ecological significance. Worm 1(2):98–106. https://doi.org/10.4161/worm.21086
Walker JE, Arizmendi JM, Dupuis A, Fearnley IM, Finel M, Medd SM, Pilkington SJ, Runswick MJ, Skehel JM (1992) Sequences of 20 subunits of NADH: Ubiquinone oxidoreductase from bovine heart mitochondria. Application of a novel strategy for sequencing proteins using the polymerase chain reaction. J Mol Biol 226(4):1051–1072. https://doi.org/10.1016/0022-2836(92)91052-Q
Walsh MR, Castoe T, Holmes J, Packer M, Biles K, Walsh M, Munch SB, Post DM (2016) Local adaptation in transgenerational responses to predators. Proc R Soc B Biol Sci 283(1823):20152271. https://doi.org/10.1098/rspb.2015.2271
Yin J, Zhou M, Lin Z, Li QQ, Zhang Y-Y (2019) Transgenerational effects benefit offspring across diverse environments: a meta-analysis in plants and animals. Ecol Lett 22(11):1976–1986. https://doi.org/10.1111/ele.13373
Zhu A, Ibrahim JG, Love MI (2019) Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35(12):2084–2092. https://doi.org/10.1093/bioinformatics/bty895
Acknowledgements
We are grateful to Cyril Degletagne who helped in experimental works and to the Rhône-Alpes bioinformatic centre (PRABI) that has supported bioinformatics analyses. This work was financially supported by the BioEnviS research federation (FR3728) of Lyon University and the TEATIME (ANR-21-CE02-0005) grants from the French National Research Agency (ANR).
Funding
This work was financially supported by the TEATIME (ANR-21-CE02-0005) grants from the French National Research Agency (ANR).
Author information
Authors and Affiliations
Contributions
EL, SP and TL conceived the project. JT, SP and EL performed the experiments. LK performed RNA extractions and libraries. LD, JT and MH analysed phenotypic data. LD, NS and TL performed bioinformatic molecular analyses. L.D. wrote the first version of the manuscript. All authors revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
This work did not require ethical approval from a human subject or animal welfare committee.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Sebastián Ramos-Onsins.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dejeux, L., Saclier, N., Tariel-Adam, J. et al. Transcriptomic basis of within- and trans-generational predator-induced plasticity in the freshwater snail Physa acuta. Heredity (2025). https://doi.org/10.1038/s41437-025-00775-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41437-025-00775-9