Fig. 1: Assembly of NC358 using various read lengths and coverage.
From: Effect of sequence depth and length in long-read assembly of the maize inbred NC358
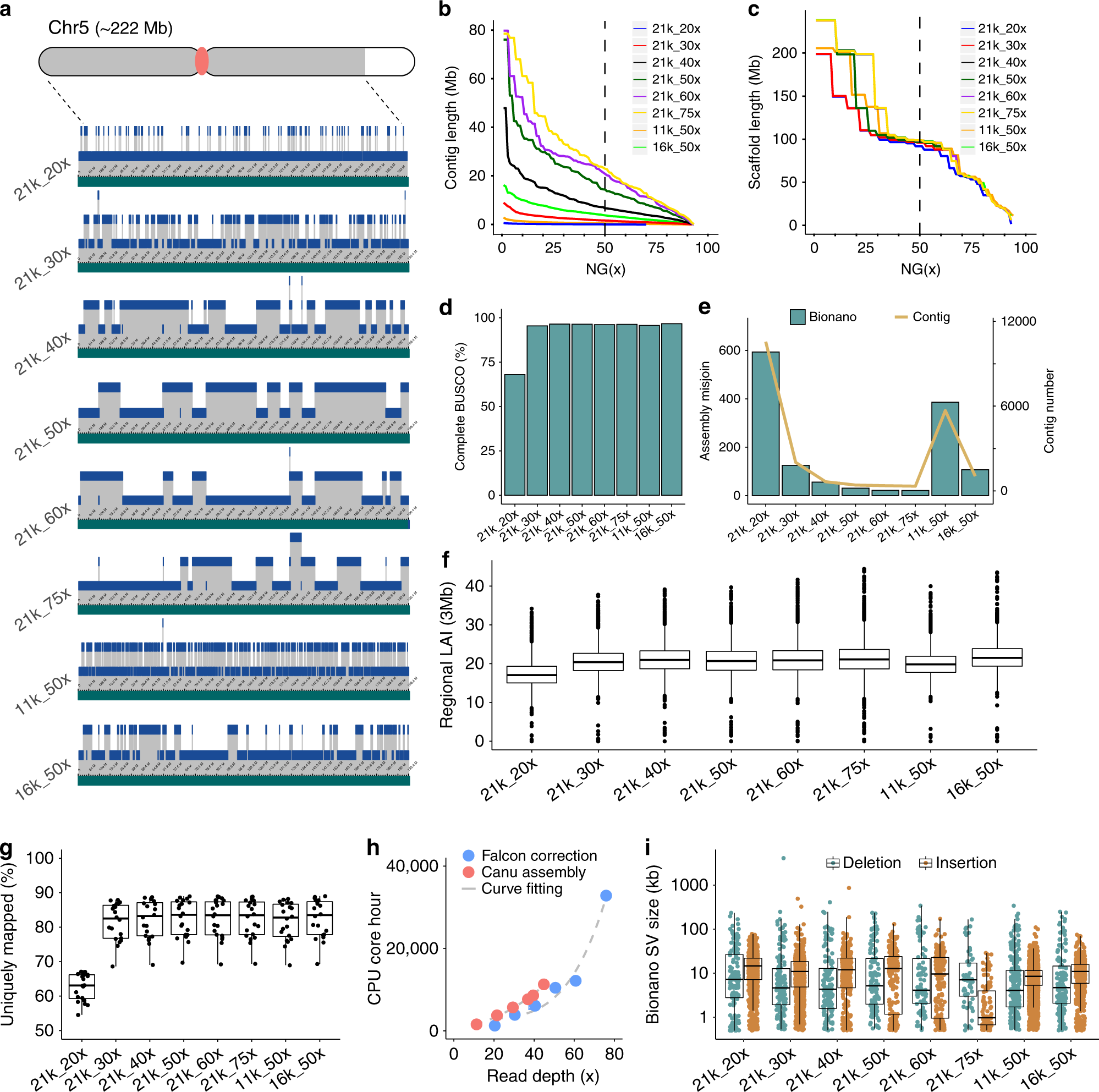
a Hybrid scaffolding using the Bionano optical map. A 199 Mb scaffold from chromosome 5 is shown. Gray areas on the chromosome cartoon represent the 199 Mb scaffold; the white area is the remaining 23 Mb scaffold in chromosome 5; the red dot is the centromere. Green tracts represent scaffolded sequences and blue tracts show the contigs that comprise this scaffold with contigs jittered across three levels. b Contig NG(x). c Scaffold NG(x). d Benchmarking Universal Single-Copy Orthologs (BUSCO) of Pilon-polished assemblies. e The number of assembly misjoins revealed by DLE-1 conflicts and the number of contigs of each assembly. f Regional LTR Assembly Index (LAI) values estimated based on 3 Mb windows with 300 kb steps. The box shows the median, upper, and lower quartiles. Whiskers indicate values ≤ 1.5× interquartile range. Outliers are plotted as dots. g Unique mapping rate of RNA-seq libraries. A total of ten tissues with two biological replicates were sequenced. Each dot represents an RNA-seq library. The box shows the median, upper, and lower quartiles. Whiskers indicate values ≤ 1.5× interquartile range. h Central processing unit (CPU) core hours required for Falcon correction and Canu assembly. i Bionano optical map inconsistency. Deletions and insertions are cases where sequences are shorter or longer than the size estimated by the optical map, respectively. Structural variations (SVs) are plotted as dots. The box shows the median, upper and lower quartiles. Whiskers indicate values ≤ 1.5× interquartile range. Source data underlying Fig. 1b, c, f, g are provided as a Source Data file.
