Fig. 1: NAP-seq accurately identifies full-length napRNAs at single-nucleotide resolution.
From: NAP-seq reveals multiple classes of structured noncoding RNAs with regulatory functions
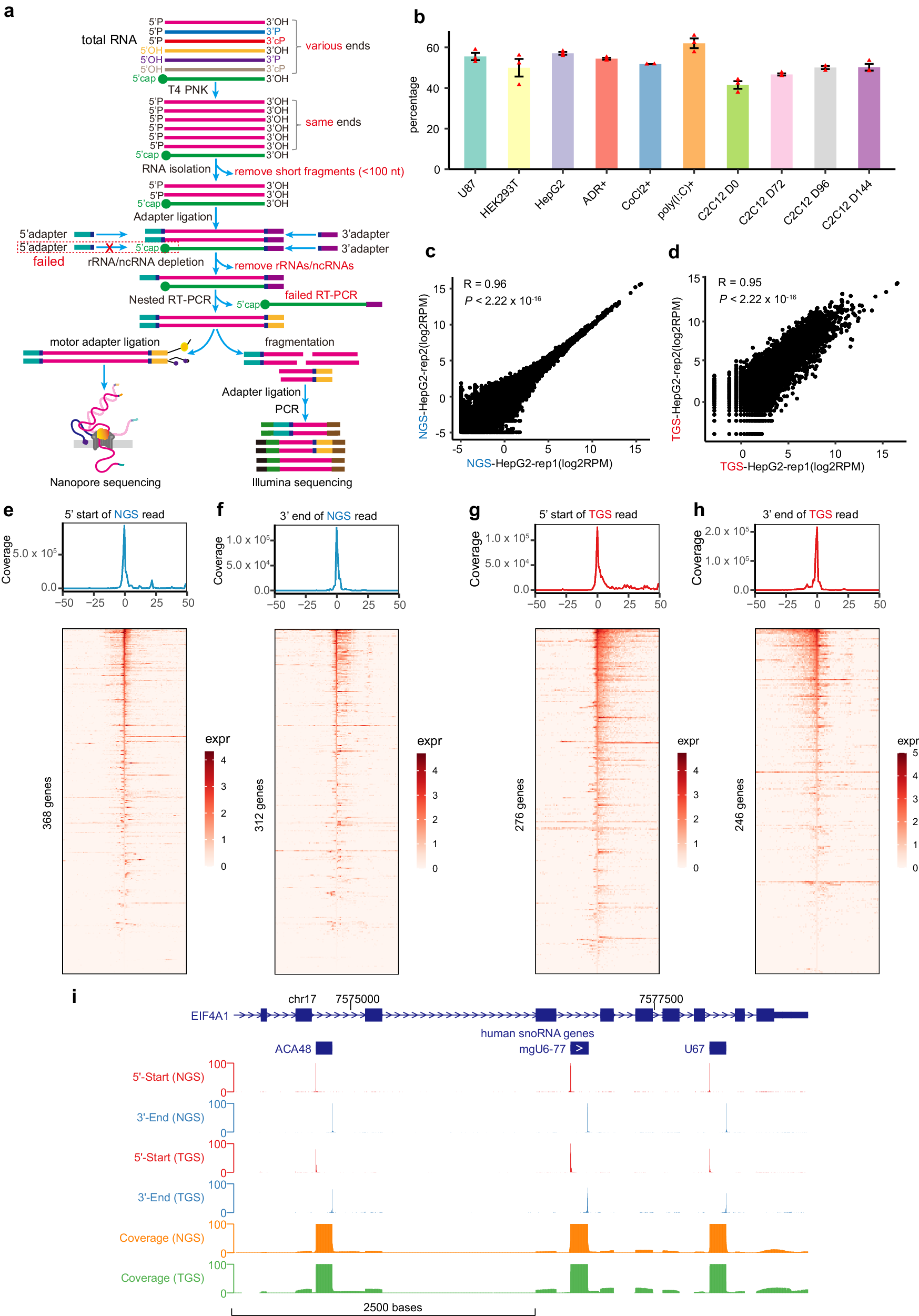
a Schematic depiction of the NAP-seq method with the construction of TGS libraries and NGS libraries. Total RNA was extracted and pre-size selection of RNA fractions was performed by removing short RNAs. b The percentage of reads with at least one specific adapter in the NAP-seq-NGS libraries in humans and mice (3 biological replicates per sample). All data are plotted as the means ± SEMs. The number of reads (RPM, reads per million) for known ncRNAs was highly reproducible between replicates with both the NAP-seq-NGS (c) and NAP-seq-TGS (d) methods. p values were calculated by two-sided Pearson’s correlation test. e, f Coincidence analysis of both terminal sites in the NAP-seq-NGS reads and those in the known ncRNAs in HepG2 cells. The x-axis shows the distance from the annotated 5’-start (e) or 3’-end (f) site in the NAP-seq-NGS read to the corresponding terminal sites in the known ncRNAs, and the y-axis shows the number of reads (RPM) within a certain distance from the 5’-start or 3’-end site. The bottom panel shows a heatmap, in which each row represents a gene that matches the terminal sites with the NAP-seq-NGS read exactly, and each column represents the expression values of genes at a specific distance. expr, expression value. g Number of coincidences between the 5’-start sites in NAP-seq-TGS reads and the 5’-start sites in known ncRNAs in HepG2 cells. h Number of coincidences between the 3’-end sites in NAP-seq-TGS reads and the 3’-end sites in known ncRNAs in HepG2 cells. i Genome Browser visualization of NAP-seq 5’-start, 3’-end and coverage signals (RPM, reads per million), which were generated from NAP-seq-NGS and NAP-seq-TGS data, in a 5000-bp region of a human snoRNA cluster (chr17:7,573,750-7,578,750).
