Fig. 1: MapUTR captures functional 3′ UTR variants in known functional motifs.
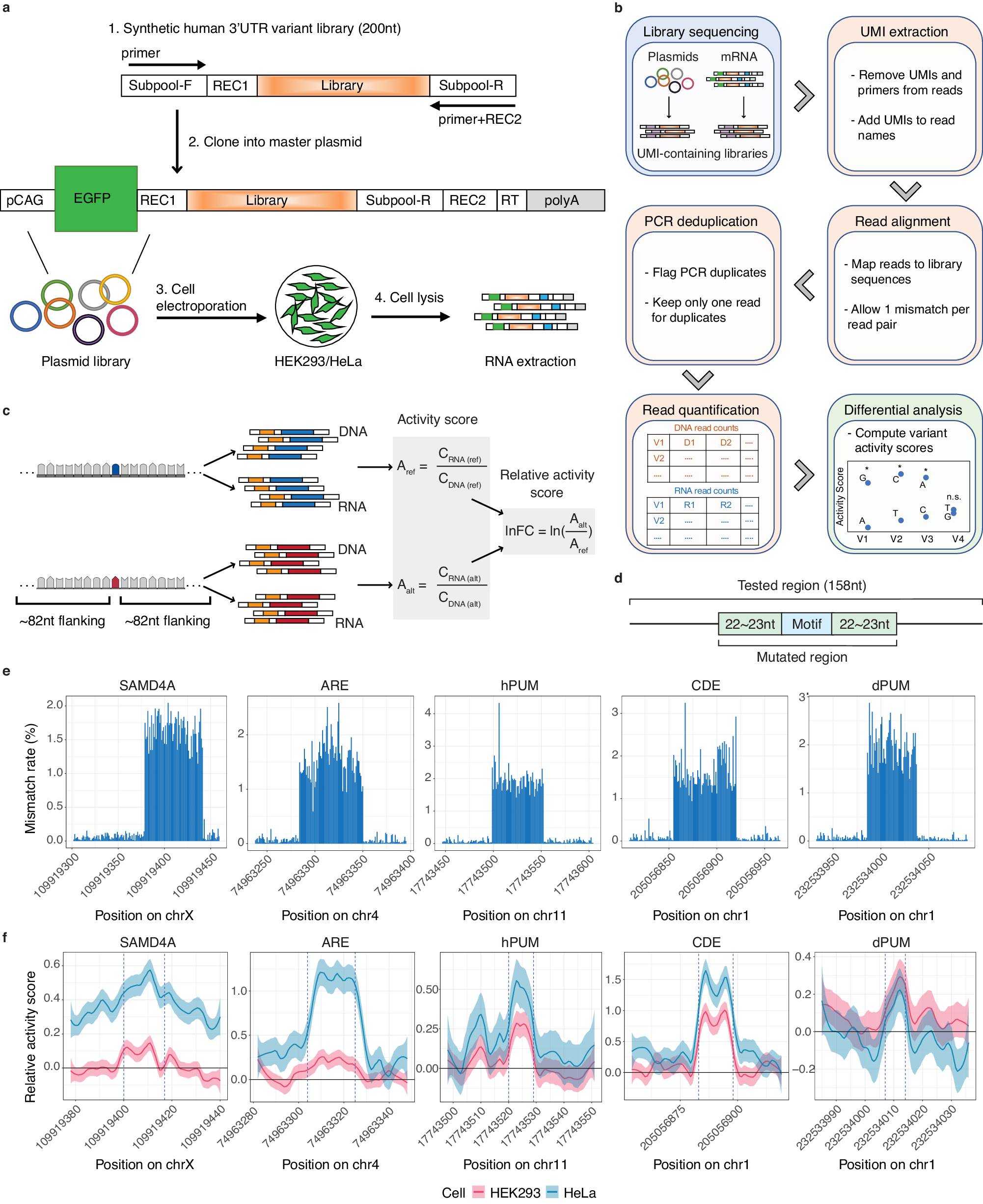
a Experimental Design of MapUTR. Subpool-F and Subpool-R represent the 15-mer primer sequences to amplify the oligo pool. REC1 and REC2 represent restriction enzyme sites. pCAG, the CMV early enhancer/chicken beta actin (CAG) promoter. EGFP, eGFP gene. Library, oligo sequences containing the mutations/motifs. RT, sequences for gene-specific reverse transcription. polyA, polyA signals. See also Supplementary Fig. 2. b Computational workflow. c Design of oligos containing rare variants from gnomAD and quantification of variant effects. CDNA (ref) represents DNA counts for the reference allele, CRNA (ref) represents RNA counts for the reference allele, CDNA (alt) represents DNA counts for the alternative allele, CRNA (alt) represents RNA counts for the alternative allele, Aref represents the activity score of the reference allele, Aalt represents the activity score of the alternative allele, and lnFC represents the relative activity score of the alternative allele compared to the reference allele. Created with BioRender.com. d Diagram of oligos with random mutations in the motif and its flanking regions (22–23 nt). e Mismatch rate (%) per position along the length of DNA sequences harboring known motifs: SAMD4A, sterile alpha motif ___domain containing 4A motif (in gene CHRDL1), ARE, AU-rich element (in gene CXCL2), hPUM, human pumilio motif (in gene MYOD1), CDE, constitutive decay element (in gene RBBP5), and dPUM, Drosophila pumilio motif (in gene SIPA1L2). f Relative activity scores of each mutated position of known motifs in HEK293 (red) and HeLa (blue) cells. Relative activity scores are averaged across the 3 tested alternative alleles per position. The 95% confidence intervals are shown as shades. e, f Source data are provided as a Source Data file.
