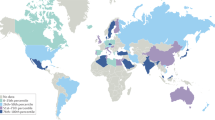
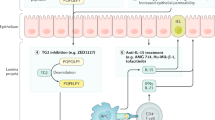
Abstract
Progress has been made in understanding coeliac disease, a relatively frequent and underappreciated immune-mediated condition that occurs in genetically predisposed individuals. However, several gaps remain in knowledge related to diagnosis and management. The gluten-free diet, currently the only available management, is not curative or universally effective (some adherent patients have ongoing duodenal injury). Unprecedented numbers of emerging therapies, including some with novel tolerogenic mechanisms, are currently being investigated in clinical trials. In March 2020, the Celiac Disease Foundation and the Society for the Study of Celiac Disease convened a consensus workshop to identify high-yield areas of research that should be prioritized. Workshop participants included leading experts in clinical practice, academia, government and pharmaceutical development, as well as representatives from patient support groups in North America. This Roadmap summarizes key advances in the field of coeliac disease and provides information on important discussions from the consensus approach to address gaps and opportunities related to the pathogenesis, diagnosis and management of coeliac disease. The morbidity of coeliac disease is often underestimated, which has led to an unmet need to improve the management of these patients. Expanded research funding is needed as coeliac disease is a potentially curable disease.
Key points
-
Coeliac disease is a common and serious medical condition that is under-recognized among the health-care provider community, government and the public.
-
This Roadmap summarizes consensus recommendations to address gaps and opportunities in pathogenesis, diagnosis and management of coeliac disease.
-
Various animal models are available to translate hypotheses generated from human studies, and progress is being made in the development of physiological coeliac epithelial models based on organoid technology.
-
Coeliac-specific serology is highly reliable for the diagnosis of coeliac disease; however, there is disagreement between experts as to the necessity of intestinal biopsy to confirm the diagnosis.
-
There is increasing need for development of programmes for proper clinical management of coeliac disease, and the number of potential therapeutic targets and clinical trials has grown exponentially over the past 15 years.
-
Increased funding for coeliac disease research is crucial to improve clinical management and facilitate development of therapies for this condition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Singh, P. et al. Global prevalence of celiac disease: systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 16, 823–836 (2018).
Lebwohl, B., Sanders, D. S. & Green, P. H. R. Coeliac disease. Lancet 391, 70–81 (2018).
King, J. A. et al. Incidence of celiac disease is increasing over time: a systematic review and meta-analysis. Am. J. Gastroenterol. 115, 507–525 (2020).
Lebwohl, B., Green, P. H. R., Söderling, J., Roelstraete, B. & Ludvigsson, J. F. Association between celiac disease and mortality risk in a swedish population. JAMA 323, 1277–1285 (2020).
Rubio-Tapia, A., Hill, I. D., Kelly, C. P., Calderwood, A. H. & Murray, J. A. ACG clinical guidelines: diagnosis and management of celiac disease. Am. J. Gastroenterol. 108, 656–676; quiz 677 (2013).
Geller, M. G. SSCD consensus workshop on celiac disease research – Marilyn’s Message March 2020. Celiac Disease Foundation https://celiac.org/about-the-foundation/featured-news/2020/03/marilyns-message-march-2020/ (2020).
Tye-Din, J. A. et al. Patient factors influencing acute gluten reactions and cytokine release in treated coeliac disease. BMC Med. 18, 362 (2020).
Sollid, L. M. et al. Update 2020: nomenclature and listing of celiac disease–relevant gluten epitopes recognized by CD4+ T cells. Immunogenetics 72, 85–88 (2020).
Kim, C.-Y., Quarsten, H., Bergseng, E., Khosla, C. & Sollid, L. M. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc. Natl Acad. Sci. USA 101, 4175–4179 (2004).
Henderson, K. N. et al. A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity 27, 23–34 (2007).
Goel, G. et al. Cytokine release and gastrointestinal symptoms after gluten challenge in celiac disease. Sci. Adv. 5, eaaw7756 (2019).
Gutierrez-Achury, J. et al. Fine mapping in the MHC region accounts for 18% additional genetic risk for celiac disease. Nat. Genet. 47, 577–578 (2015).
Kuja-Halkola, R. et al. Heritability of non-HLA genetics in coeliac disease: a population-based study in 107 000 twins. Gut 65, 1793–1798 (2016).
Marietta, E. V. & Murray, J. A. Animal models to study gluten sensitivity. Semin. Immunopathol. 34, 497–511 (2012).
de Kauwe, A. L. et al. Resistance to celiac disease in humanized HLA-DR3-DQ2-Transgenic mice expressing specific anti-gliadin CD4+ T cells. J. Immunol. 182, 7440–7450 (2009).
Abadie, V. et al. IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature 578, 600–604 (2020).
Caminero, A. et al. Duodenal bacterial proteolytic activity determines sensitivity to dietary antigen through protease-activated receptor-2. Nat. Commun. 10, 1198 (2019).
Caminero, A. et al. Lactobacilli degrade wheat amylase trypsin inhibitors to reduce intestinal dysfunction induced by immunogenic wheat products. Gastroenterology 156, 2266–2280 (2019).
Lamas, B. et al. Aryl hydrocarbon receptor ligand production by the gut microbiota is decreased in celiac disease leading to intestinal inflammation. Sci. Transl. Med. 12, eaba0624 (2020).
Bouziat, R. et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 356, 44–50 (2017).
Dieterich, W., Neurath, M. F. & Zopf, Y. Intestinal ex vivo organoid culture reveals altered programmed crypt stem cells in patients with celiac disease. Sci. Rep. 10, 3535 (2020).
Freire, R. et al. Human gut derived-organoids provide model to study gluten response and effects of microbiota-derived molecules in celiac disease. Sci. Rep. 9, 7029 (2019).
Serena, G. et al. Intestinal epithelium modulates macrophage response to gliadin in celiac disease. Front. Nutr. 6, 167 (2019).
Fujii, M., Clevers, H. & Sato, T. Modeling human digestive diseases with CRISPR–Cas9-modified organoids. Gastroenterology 156, 562–576 (2019).
Sarvestani, S. K. et al. Induced organoids derived from patients with ulcerative colitis recapitulate colitic reactivity. Nat. Commun. 12, 262 (2021).
Blansky, B. A. et al. Lack of follow-up of pediatric patients with celiac disease. Clin. Gastroenterol. Hepatol. 17, 2603–2604 (2019).
Hayter, S. M. & Cook, M. C. Updated assessment of the prevalence, spectrum and case definition of autoimmune disease. Autoimmun. Rev. 11, 754–765 (2012).
Christophersen, A. et al. Distinct phenotype of CD4+ T cells driving celiac disease identified in multiple autoimmune conditions. Nat. Med. 25, 734–737 (2019).
Leonard, M. M. et al. Evaluating responses to gluten challenge: a randomized, double-blind, 2-dose gluten challenge trial. Gastroenterology 160, 720–733.e8 (2021).
Verdu, E. F. & Danska, J. S. Common ground: shared risk factors for type 1 diabetes and celiac disease. Nat. Immunol. 19, 685–695 (2018).
Andrén Aronsson, C. et al. Association of gluten intake during the first 5 years of life with incidence of celiac disease autoimmunity and celiac disease among children at increased risk. JAMA 322, 514–523 (2019).
Lindfors, K. et al. Metagenomics of the faecal virome indicate a cumulative effect of enterovirus and gluten amount on the risk of coeliac disease autoimmunity in genetically at risk children: the TEDDY study. Gut 69, 1416–1422 (2020).
Bibbins-Domingo, K. et al. Screening for celiac disease: US preventive services task force recommendation statement. JAMA 317, 1252–1257 (2017).
Hujoel, I. A. et al. Natural history and clinical detection of undiagnosed coeliac disease in a North American community. Aliment. Pharmacol. Ther. 47, 1358–1366 (2018).
Stahl, M. G. et al. Mass screening for celiac disease: the autoimmunity screening for kids study. Am. J. Gastroenterol. 116, 180–187 (2021).
Husby, S. et al. European society paediatric gastroenterology, hepatology and nutrition guidelines for diagnosing coeliac disease 2020. J. Pediatr. Gastroenterol. Nutr. 70, 141–156 (2020).
Riznik, P. et al. The use of biopsy and ‘no-biopsy’ approach for diagnosing paediatric coeliac disease in the central european region. Gastroenterol. Res. Pract. 2019, 9370397 (2019).
Rees, C. J. et al. Restarting gastrointestinal endoscopy in the deceleration and early recovery phases of COVID-19 pandemic: guidance from the British Society of Gastroenterology. Clin. Med. 20, 352–358 (2020).
Leffler, D. et al. Kinetics of the histological, serological and symptomatic responses to gluten challenge in adults with coeliac disease. Gut 62, 996–1004 (2013).
Penny, H. A., Baggus, E. M. R., Rej, A., Snowden, J. A. & Sanders, D. S. Non-responsive coeliac disease: a comprehensive review from the NHS england national centre for refractory coeliac disease. Nutrients 12, 216 (2020).
Sarna, V. K. et al. HLA-DQ–gluten tetramer blood test accurately identifies patients with and without celiac disease in absence of gluten consumption. Gastroenterology 154, 886–896.e6 (2018).
Anderson, R. P. et al. Whole blood interleukin-2 release test to detect and characterize rare circulating gluten-specific T cell responses in coeliac disease. Clin. Exp. Immunol. 204, 321–334 (2021).
van de Kamer, J. H., Weijers, H. A. & Dicke, W. K. An investigation inthe the injurious constituents of wheat in connection with their action on patients with coeliac disease. Acta Paediatr. 42, 223–231 (1953).
Sollid, L. et al. Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J. Exp. Med. 169, 345–350 (1989).
Dieterich, W. et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 3, 797–801 (1997).
Codex Alimentarius Commission. Codex standard for foods for special dietary use for persons intolerant to gluten. CSX 118-1979 (WHO, 2008).
Abu-Janb, N. & Jaana, M. Facilitators and barriers to adherence to gluten-free diet among adults with celiac disease: a systematic review. J. Hum. Nutr. Diet. 33, 786–810 (2020).
Silvester, J. A. et al. Most patients with celiac disease on gluten-free diets consume measurable amounts of gluten. Gastroenterology 158, 1497–1499.e1 (2020).
O’Leary, C. et al. Celiac disease and the transition from childhood to adulthood: a 28-year follow-up. Am. J. Gastroenterol. 99, 2437–2441 (2004).
Lebwohl, B., Murray, J. A., Rubio-Tapia, A., Green, P. H. R. & Ludvigsson, J. F. Predictors of persistent villous atrophy in coeliac disease: a population-based study. Aliment. Pharmacol. Ther. 39, 488–495 (2014).
Choung, R. S. et al. Prevalence and morbidity of undiagnosed celiac disease from a community-based study. Gastroenterology 152, 830–839.e5 (2017).
Mearns, E. S. et al. Systematic literature review of the economic burden of celiac disease. Pharmacoeconomics 37, 45–61 (2019).
Kivelä, L. et al. Current and emerging therapies for coeliac disease. Nat. Rev. Gastroenterol. Hepatol. 18, 181–195 (2021).
Schuppan, D. et al. A randomized trial of a transglutaminase 2 inhibitor for celiac disease. N. Engl. J. Med. 385, 35–45 (2021).
Clerx, E., Kupfer, S. S. & Leffler, D. A. Disparities among gastrointestinal disorders in research funding from the national institutes of health. Gastroenterology 153, 877–880 (2017).
Liu, E. et al. Risk of pediatric celiac disease according to HLA haplotype and country. N. Engl. J. Med. 371, 42–49 (2014).
Rubio-Tapia, A., Ludvigsson, J. F., Brantner, T. L., Murray, J. A. & Everhart, J. E. The prevalence of celiac disease in the United States. Am. J. Gastroenterol. 107, 1538–1544 (2012).
Comino, I. et al. Monitoring of gluten-free diet compliance in celiac patients by assessment of gliadin 33-mer equivalent epitopes in feces. Am. J. Clin. Nutr. 95, 670–677 (2012).
Silvester, J. A. et al. Exposure sources, amounts and time course of gluten ingestion and excretion in patients with coeliac disease on a gluten-free diet. Aliment. Pharmacol. Ther. 52, 1469–1479 (2020).
TEDDY Study Group. The environmental determinants of diabetes in the young (TEDDY) study: study design. Pediatr. Diabetes 8, 286–298 (2007).
Getts, D. R., Shea, L. D., Miller, S. D. & King, N. J. C. Harnessing nanoparticles for immune modulation. Trends Immunol. 36, 419–427 (2015).
O’Leary, M., Krailo, M., Anderson, J. R. & Reaman, G. H. Progress in childhood cancer: 50 years of research collaboration, a report from the Children’s Oncology Group. Semin. Oncol. 35, 484–493 (2008).
PALISADE Group of Clinical Investigators. AR101 oral immunotherapy for peanut allergy. N. Engl. J. Med. 379, 1991–2001 (2018).
Egberg, M. D., Kappelman, M. D. & Gulati, A. S. Improving care in pediatric inflammatory bowel disease. Gastroenterol. Clin. North. Am. 47, 909–919 (2018).
Mogayzel, P. J., Dunitz, J., Marrow, L. C. & Hazle, L. A. Improving chronic care delivery and outcomes: the impact of the cystic fibrosis Care Center Network. BMJ Qual. Saf. 23, 3–8 (2014).
Osborne, T. B. Our present knowledge of plant proteins. Science 28, 417–427 (1908).
Rej, A., Aziz, I. & Sanders, D. S. Breaking bread! Proc. Nutr. Soc. 78, 118–125 (2019).
Asri, N., Rostami-Nejad, M., Anderson, R. P. & Rostami, K. The gluten gene: unlocking the understanding of gluten sensitivity and intolerance. Appl. Clin. Genet. 14, 37–50 (2021).
Hardy, M. Y. et al. Ingestion of oats and barley in patients with celiac disease mobilizes cross-reactive T cells activated by avenin peptides and immuno-dominant hordein peptides. J. Autoimmun. 56, 56–65 (2015).
Solheim, B. G. et al. HLA antigens in dermatitis herpetiformis and coeliac disease. Tissue Antigens 7, 57–59 (1976).
Sallese, M., Lopetuso, L. R., Efthymakis, K. & Neri, M. Beyond the HLA genes in gluten-related disorders. Front. Nutr. 7, 1–7 (2020).
Ju, J. M., Marietta, E. V. & Murray, J. A. Generating transgenic mouse models for studying celiac disease. Methods Mol. Biol. 1326, 23–33 (2015).
Lundin, K. E. et al. Gliadin-specific, HLA-DQ(alpha 1*0501,beta 1*0201) restricted T cells isolated from the small intestinal mucosa of celiac disease patients. J. Exp. Med. 178, 187–196 (1993).
Chen, X. et al. Structural basis for antigen recognition by transglutaminase 2-specific autoantibodies in celiac disease. J. Biol. Chem. 290, 21365–21375 (2015).
Mayassi, T. & Jabri, B. Human intraepithelial lymphocytes. Mucosal Immunol. 11, 1281–1289 (2018).
Tye-Din, J. A. et al. Elevated serum interleukin-2 after gluten correlates with symptoms and is a potential diagnostic biomarker for coeliac disease. Aliment. Pharmacol. Ther. 50, 901–910 (2019).
Caminero, A. & Verdu, E. F. Celiac disease: should we care about microbes? Am. J. Physiol. - Gastrointest. Liver Physiol. 317, G161–G170 (2019).
Shiner, M. Duodenal biopsy. Lancet 270, 17–19 (1956).
Cichewicz, A. B. et al. Diagnosis and treatment patterns in celiac disease. Dig. Dis. Sci. 64, 2095–2106 (2019).
Anderson, C. M. Histological changes in the duodenal mucosa in coeliac disease: Reversibility during treatment with a wheat gluten free diet. Arch. Dis. Child. 35, 419–427 (1960).
Silvester, J. A. et al. Tests for serum transglutaminase and endomysial antibodies do not detect most patients with celiac disease and persistent villous atrophy on gluten-free diets: a meta-analysis. Gastroenterology 153, 689–701.e1 (2017).
Lähdeaho, M. L. et al. Safety and efficacy of AMG 714 in adults with coeliac disease exposed to gluten challenge: a phase 2a, randomised, double-blind, placebo-controlled study. Lancet Gastroenterol. Hepatol. 4, 948–959 (2019).
Daveson, A. J. M. et al. Baseline quantitative histology in therapeutics trials reveals villus atrophy in most patients with coeliac disease who appear well controlled on gluten-free diet. GastroHep 2, 22–30 (2020).
Morón, B. et al. Toward the assessment of food toxicity for celiac patients: characterization of monoclonal antibodies to a main immunogenic gluten peptide. PLoS ONE 3, e2294 (2008).
Shalimar et al. Effect of addition of short course of prednisolone to gluten-free diet on mucosal epithelial cell regeneration and apoptosis in celiac disease: a pilot randomized controlled trial. Dig. Dis. Sci. 57, 3116–3125 (2012).
Lionetti, E. et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N. Engl. J. Med. 371, 1295–1303 (2014).
Gottlieb, K., Dawson, J., Hussain, F. & Murray, J. A. Development of drugs for celiac disease: review of endpoints for phase 2 and 3 trials. Gastroenterol. Rep. 3, 91–102 (2015).
Leffler, D. et al. Development of celiac disease therapeutics: report of the third gastroenterology regulatory endpoints and advancement of therapeutics workshop. Gastroenterology 151, 407–411 (2016).
Vriezinga, S. L. et al. Randomized feeding intervention in infants at high risk for celiac disease. N. Engl. J. Med. 371, 1304–1315 (2014).
Acknowledgements
M.I.P.-S. received an Innovation Grant from CCC, Medicine Internal Career Research Award, Division of Gastroenterology AFP grant, and Farncombe Family Digestive Health Research Institute Award. J.A.S. is supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH under award number K23DK119584. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Canadian Institutes of Health Research. E.F.V is funded by a CIHR grant 168840 and holds a Canada Research Chair. R.P.A. declares no funding. The authors thank the hospitality of the Faculty House at Columbia University and Implementation Committee, C. Beckman and K. Cervenka. The authors thank the organizing committee, P. H. R. Green and J. A. Murray, the scientific committee P. H. R. Green (Society for the Study of Celiac disease (SSCD) Past President), C.P.K. (SSCD President), B.L. (SSCD President Elect), J. A. Murray. (SSCD Past President) and E.F.V. (SSCD Past President). Special thanks for the contribution of other Consensus Workshop Faculty: D. Adams (Vanderbilt Center Celiac Clinic, USA); A. Alaedini, G. Bhagat, S. Krishnareddy, N. R. Reilly, A. R. Lee (Columbia University Celiac Disease Center, USA); S. Moleski (Jefferson Celiac Center, USA); B. Tycko (John Threurer Cancer Center, USA); M. J. Blaser (Rutgers University, USA); S. P. Burke, C. Elson; R. S. Chuong and L. Harris (Mayo Clinic. USA); B. Jabri, C. Semrad and R. Verma (University of Chicago, USA); M. Leonard and A. Fasano (Center for Celiac Research and Treatment at Massachussets General Hospital, USA); E. Liu (Children’s Hospital Colorado, USA); A. Cartee (University of Michigan, USA); E. Charles (University of Alabama, USA); A. Rothermel (NIAID, NIH, DHHS); M. Geller, D. Ceizler and R. English (Celiac Disease Foundation); F. Leon (Provention Bio) and A. Sapone (Takeda Pharmaceuticals). Only participants of the workshop included as authors were involved in the writing of this Roadmap Review.
Author information
Authors and Affiliations
Contributions
All authors participated in the conception, writing and editing of the article.
Corresponding author
Ethics declarations
Competing interests
B.L. has consulted for Takeda and Anokion. D.A.L. receives salary support from Takeda Pharmaceuticals unrelated to this manuscript. M.I.P.-S. has consulted for Takeda and Lupin unrelated to this manuscript. J.A.S. has consulted for Takeda Pharmaceuticals and received grant funding from Glutenostics, Milky Way Life Sciences, the Celiac Disease Foundation and BEYOND Celiac. E.F.V. received grant funding from Biocodex Foundation and Gilead unrelated to this manuscript. R.P.A. has served as consultant for Takeda, GSK, Anokion, Allero Therapeutics, TregTherapeutics and EVOQ Therapeutics. R.P.A. is founder and shareholder of Novoviah Pharmaceuticals and is the inventor of patents relating to the diagnosis and treatment of coeliac disease. The remaining authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks Stefano Guandalini, Knut Lundin, David Sanders and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Celiac Disease Foundation: https://celiac.org/
Coeliac disease centres: https://celiac.org/celiacdiseasecentersandprograms/
ImproveCareNow: https://www.improvecarenow.org/
SEER database: https://seer.cancer.gov/
Surveillance Research Program: https://surveillance.cancer.gov/
Society for the Study of Celiac Disease: https://www.theceliacsociety.org/
TrialNet: https://www.trialnet.org/
Rights and permissions
About this article
Cite this article
Pinto-Sanchez, M.I., Silvester, J.A., Lebwohl, B. et al. Society for the Study of Celiac Disease position statement on gaps and opportunities in coeliac disease. Nat Rev Gastroenterol Hepatol 18, 875–884 (2021). https://doi.org/10.1038/s41575-021-00511-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-021-00511-8
This article is cited by
-
Efficacy and safety of gut microbiota-based therapies in autoimmune and rheumatic diseases: a systematic review and meta-analysis of 80 randomized controlled trials
BMC Medicine (2024)
-
A machine learning tool for early identification of celiac disease autoimmunity
Scientific Reports (2024)
-
Tolerance-inducing therapies in coeliac disease — mechanisms, progress and future directions
Nature Reviews Gastroenterology & Hepatology (2024)
-
A human autoimmune organoid model reveals IL-7 function in coeliac disease
Nature (2024)
-
Transcriptome profile and immune infiltrated landscape revealed a novel role of γδT cells in mediating pyroptosis in celiac disease
Journal of Translational Medicine (2023)