Fig. 1: Overview of experiments and data types.
From: A large-scale binding and functional map of human RNA-binding proteins
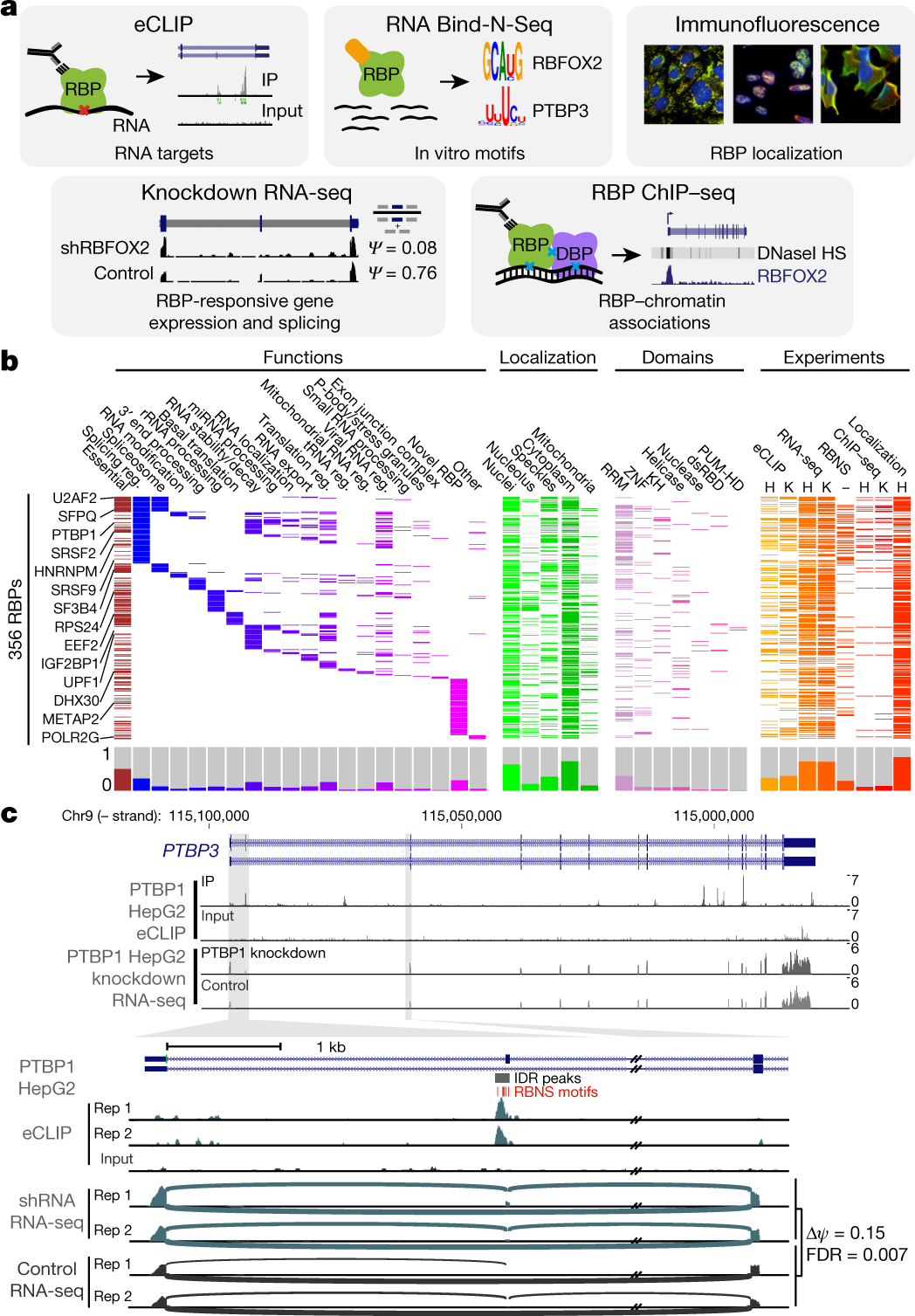
a, The five assays performed to characterize RBPs. b, Three hundred and fifty-six RBPs profiled by at least one ENCODE experiment (orange or red) with localization by immunofluorescence (green), essential genes from CRISPR screening (maroon), manually annotated RBP functions (blue or purple), and annotated protein domains (pink; RRM, KH, zinc finger, RNA helicase, RNase, double-stranded RNA binding (dsRBD), and pumilio/FBF ___domain (PUM-HD)). Histograms for each category are shown at bottom. c, Combinatorial expression and splicing regulation of PTBP3. Tracks indicate eCLIP and RNA-seq read density (reads per million). Tracks are shown for replicate 1; eCLIP and KD–RNA-seq were performed in biological duplicate with similar results. Bottom, alternatively spliced exon 2, with lines indicating junction-spanning reads and indicated per cent spliced in (ψ). Boxes indicate reproducible (by IDR) PTBP1 peaks, with red boxes indicating RBNS motifs for the PTB family member PTBP3 located within (or up to 50 bases upstream of) peaks.
