Fig. 1: Overview of Sniffles2.
From: Detection of mosaic and population-level structural variants with Sniffles2
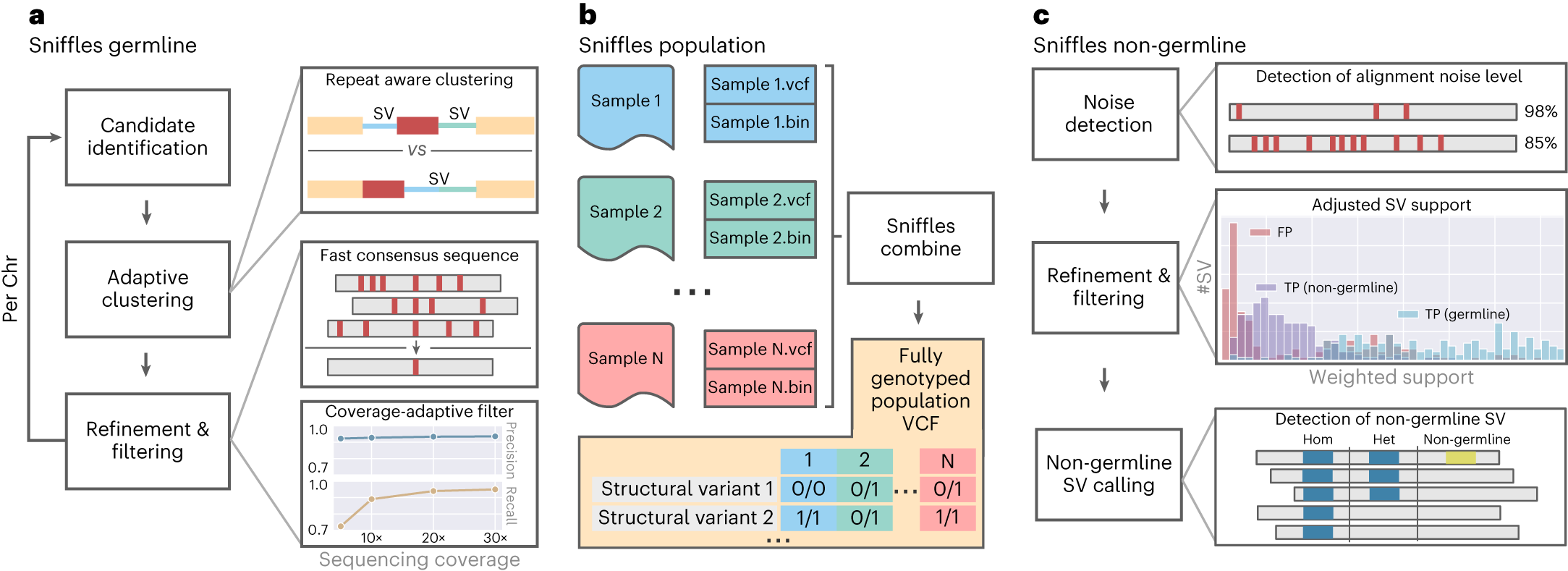
a, For Sniffles2, we implemented a repeat aware clustering coupled with a fast consensus sequence and coverage-adaptive filtering to improve accuracy of the germline SV calls. b, One key limitation of current SV calling is the generation of fully genotyped population VCF. Sniffles2 implements a concept similar to a gVCF file where single-sample calling is done only once, which reduces runtime multiple-fold. c, Mosaic SV detection is enabled by improved detection and filtering of low VAF SVs (by default, 5–20%) across a bulk sample. This is enabled over additional noise detection methodology as well as refinement and filtering approaches that we developed.
