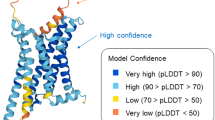
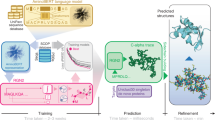
Abstract
Artificial intelligence-driven advances in protein structure prediction in recent years have raised the question: has the protein structure-prediction problem been solved? Here, with a focus on nonglobular proteins, we highlight the many strengths and potential weaknesses of DeepMind’s AlphaFold2 in the context of its biological and therapeutic applications. We summarize the subtleties associated with evaluation of AlphaFold2 model quality and reliability using the predicted local distance difference test (pLDDT) and predicted aligned error (PAE) values. We highlight various classes of proteins that AlphaFold2 can be applied to and the caveats involved. Concrete examples of how AlphaFold2 models can be integrated with experimental data in the form of small-angle X-ray scattering (SAXS), solution NMR, cryo-electron microscopy (cryo-EM) and X-ray diffraction are discussed. Finally, we highlight the need to move beyond structure prediction of rigid, static structural snapshots toward conformational ensembles and alternate biologically relevant states. The overarching theme is that careful consideration is due when using AlphaFold2-generated models to generate testable hypotheses and structural models, rather than treating predicted models as de facto ground truth structures.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
PyMOL sessions containing comparisons of AlphaFold models (extracted from the literature or the AlphaFold database) compared with experimental structures together with python script used to color code structures based on pLDDT values are freely available at https://github.com/mcshanlab/AlphaFold_Models_Agarwal_McShan.
References
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Bertoline, L. M. F., Lima, A. N., Krieger, J. E. & Teixeira, S. K. Before and after AlphaFold2: an overview of protein structure prediction. Front. Bioinform. 3, 1120370 (2023).
Perrakis, A. & Sixma, T. K. AI revolutions in biology. EMBO Rep. 22, e54046 (2021).
Bouatta, N., Sorger, P. & AlQuraishi, M. Protein structure prediction by AlphaFold2: are attention and symmetries all you need? Acta Crystallogr. D 77, 982–991 (2021).
Tunyasuvunakool, K. et al. Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Varadi, M. et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 52, D368–D375 (2024).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Ahdritz, G. et al. OpenFold: retraining AlphaFold2 yields new insights into its learning mechanisms and capacity for generalization. Nat. Methods https://doi.org/10.1038/s41592-024-02272-z (2024).
Chen, S.-J. et al. Protein folds vs. protein folding: differing questions, different challenges. Proc. Natl Acad. Sci. USA 120, e2214423119 (2023).
Skolnick, J., Gao, M., Zhou, H. & Singh, S. AlphaFold 2: why it works and its implications for understanding the relationships of protein sequence, structure, and function. J. Chem. Inf. Model. 61, 4827–4831 (2021).
Outeiral, C., Nissley, D. A. & Deane, C. M. Current structure predictors are not learning the physics of protein folding. Bioinformatics 38, 1881–1887 (2022).
Alford, R. F. et al. The Rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theory Comput. 13, 3031–3048 (2017).
Roney, J. P. & Ovchinnikov, S. State-of-the-art estimation of protein model accuracy using AlphaFold. Phys. Rev. Lett. 129, 238101 (2022).
Laurents, D. V. AlphaFold 2 and NMR spectroscopy: partners to understand protein structure, dynamics and function. Front. Mol. Biosci. 9, 906437 (2022).
Chakravarty, D. & Porter, L. L. AlphaFold2 fails to predict protein fold switching. Protein Sci. 31, e4353 (2022).
Akdel, M. et al. A structural biology community assessment of AlphaFold2 applications. Nat. Struct. Mol. Biol. 29, 1056–1067 (2022).
Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at bioRxiv https://doi.org/10.1101/2021.10.04.463034 (2022).
Gao, M., Nakajima An, D., Parks, J. M. & Skolnick, J. AF2Complex predicts direct physical interactions in multimeric proteins with deep learning. Nat. Commun. 13, 1744 (2022).
Mariani, V., Biasini, M., Barbato, A. & Schwede, T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 29, 2722–2728 (2013).
Oeffner, R. D. et al. Putting AlphaFold models to work with phenix.process_predicted_model and ISOLDE. Acta Crystallogr. D 78, 1303–1314 (2022).
Tsaban, T. et al. Harnessing protein folding neural networks for peptide–protein docking. Nat. Commun. 13, 176 (2022).
McDonald, E. F., Jones, T., Plate, L., Meiler, J. & Gulsevin, A. Benchmarking AlphaFold2 on peptide structure prediction. Structure 31, 111–119 (2023).
Mikhaylov, V. et al. Accurate modeling of peptide–MHC structures with AlphaFold. Structure 32, 228–241 (2024).
Alderson, T. R., Pritišanac, I., Kolarić, Đ., Moses, A. M. & Forman-Kay, J. D. Systematic identification of conditionally folded intrinsically disordered regions by AlphaFold2. Proc. Natl Acad. Sci. USA 120, e2304302120 (2023).
Fowler, N. J. & Williamson, M. P. The accuracy of protein structures in solution determined by AlphaFold and NMR. Structure 30, 925–933 (2022).
Zweckstetter, M. NMR hawk‐eyed view of AlphaFold2 structures. Protein Sci. 30, 2333–2337 (2021).
Tejero, R., Huang, Y. J., Ramelot, T. A. & Montelione, G. T. AlphaFold models of small proteins rival the accuracy of solution NMR Structures. Front. Mol. Biosci. 9, 877000 (2022).
Thornton, J. M., Laskowski, R. A. & Borkakoti, N. AlphaFold heralds a data-driven revolution in biology and medicine. Nat. Med. 27, 1666–1669 (2021).
Saldaño, T. et al. Impact of protein conformational diversity on AlphaFold predictions. Bioinformatics 38, 2742–2748 (2022).
Hekkelman, M. L., de Vries, I., Joosten, R. P. & Perrakis, A. AlphaFill: enriching AlphaFold models with ligands and cofactors. Nat. Methods 20, 205–213 (2023).
Karelina, M., Noh, J. J. & Dror, R. O. How accurately can one predict drug binding modes using AlphaFold models? eLife 12, RP89386 (2023).
Diwan, G. D., Gonzalez-Sanchez, J. C., Apic, G. & Russell, R. B. Next generation protein structure predictions and genetic variant interpretation. J. Mol. Biol. 433, 167180 (2021).
David, A., Islam, S., Tankhilevich, E. & Sternberg, M. J. E. The AlphaFold database of protein structures: a biologist’s guide. J. Mol. Biol. 434, 167336 (2022).
Jambrich, M. A., Tusnady, G. E. & Dobson, L. How AlphaFold2 shaped the structural coverage of the human transmembrane proteome. Sci. Rep. 13, 20283 (2023).
Hegedűs, T., Geisler, M., Lukács, G. L. & Farkas, B. Ins and outs of AlphaFold2 transmembrane protein structure predictions. Cell. Mol. Life Sci. 79, 73 (2022).
Topitsch, A., Schwede, T. & Pereira, J. Outer membrane β-barrel structure prediction through the lens of AlphaFold2. Proteins 92, 3–14 (2024).
Azzaz, F., Yahi, N., Chahinian, H. & Fantini, J. The epigenetic dimension of protein structure is an intrinsic weakness of the AlphaFold program. Biomolecules 12, 1527 (2022).
Kishi, K. E. et al. Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine. Cell 185, 672–689 (2022).
Ruff, K. M. & Pappu, R. V. AlphaFold and implications for intrinsically disordered proteins. J. Mol. Biol. 433, 167208 (2021).
Wilson, C. J., Choy, W.-Y. & Karttunen, M. AlphaFold2: a role for disordered protein/region prediction? Int. J. Mol. Sci. 23, 4591 (2022).
Piovesan, D., Monzon, A. M. & Tosatto, S. C. E. Intrinsic protein disorder and conditional folding in AlphaFoldDB. Protein Sci. 31, e4466 (2022).
Lane, T. J. Protein structure prediction has reached the single-structure frontier. Nat. Methods 20, 170–173 (2023).
Del Alamo, D., Sala, D., Mchaourab, H. S. & Meiler, J. Sampling alternative conformational states of transporters and receptors with AlphaFold2. eLife 11, e75751 (2022).
Wayment-Steele, H. K. et al. Predicting multiple conformations via sequence clustering and AlphaFold2. Nature 625, 832–839 (2024).
Zhao, B., Ghadermarzi, S. & Kurgan, L. Comparative evaluation of AlphaFold2 and disorder predictors for prediction of intrinsic disorder, disorder content and fully disordered proteins. Comput. Struct. Biotechnol. J. 21, 3248–3258 (2023).
Buel, G. R. & Walters, K. J. Can AlphaFold2 predict the impact of missense mutations on structure? Nat. Struct. Mol. Biol. 29, 1–2 (2022).
Pak, M. A. et al. Using AlphaFold to predict the impact of single mutations on protein stability and function. PLoS ONE 18, e0282689 (2023).
Cheng, J. et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 381, eadg7492 (2023).
Moffat, L., Greener, J. G. & Jones, D. T. Using AlphaFold for rapid and accurate fixed backbone protein design. Preprint at bioRxiv https://doi.org/10.1101/2021.08.24.457549 (2021).
Goverde, C. A., Wolf, B., Khakzad, H., Rosset, S. & Correia, B. E. De novo protein design by inversion of the AlphaFold structure prediction network. Protein Sci. 32, e4653 (2023).
Yin, R., Feng, B. Y., Varshney, A. & Pierce, B. G. Benchmarking AlphaFold for protein complex modeling reveals accuracy determinants. Protein Sci. 31, e4379 (2022).
Yin, R. & Pierce, B. G. Evaluation of AlphaFold antibody–antigen modeling with implications for improving predictive accuracy. Protein Sci. 33, e4865 (2024).
Bryant, P., Pozzati, G. & Elofsson, A. Improved prediction of protein–protein interactions using AlphaFold2. Nat. Commun. 13, 1265 (2022).
Jeppesen, M. & André, I. Accurate prediction of protein assembly structure by combining AlphaFold and symmetrical docking. Nat. Commun. 14, 8283 (2023).
Pinheiro, F., Santos, J. & Ventura, S. AlphaFold and the amyloid landscape. J. Mol. Biol. 433, 167059 (2021).
Binder, J. L. et al. AlphaFold Illuminates half of the dark human proteins. Curr. Opin. Struct. Biol. 74, 102372 (2022).
Terwilliger, T. C. et al. Improved AlphaFold modeling with implicit experimental information. Nat. Methods 19, 1376–1382 (2022).
Terwilliger, T. C. et al. AlphaFold predictions are valuable hypotheses and accelerate but do not replace experimental structure determination. Nat. Methods 21, 110–116 (2024).
McCafferty, C. L., Pennington, E. L., Papoulas, O., Taylor, D. W. & Marcotte, E. M. Does AlphaFold2 model proteins’ intracellular conformations? An experimental test using cross-linking mass spectrometry of endogenous ciliary proteins. Commun. Biol. 6, 421 (2023).
Motmaen, A. et al. Peptide-binding specificity prediction using fine-tuned protein structure prediction networks. Proc. Natl Acad. Sci. USA 120, e2216697120 (2023).
Jussupow, A. & Kaila, V. R. I. Effective molecular dynamics from neural network-based structure prediction models. J. Chem. Theory Comput. 19, 1965–1975 (2023).
Guo, H.-B. et al. AlphaFold2 models indicate that protein sequence determines both structure and dynamics. Sci. Rep. 12, 10696 (2022).
Carugo, O. pLDDT values in AlphaFold2 protein models are unrelated to globular protein local flexibility. Crystals 13, 1560 (2023).
Zhu, W., Shenoy, A., Kundrotas, P. & Elofsson, A. Evaluation of AlphaFold-Multimer prediction on multi-chain protein complexes. Bioinformatics 39, btad424 (2023).
Fontana, P. et al. Structure of cytoplasmic ring of nuclear pore complex by integrative cryo-EM and AlphaFold. Science 376, eabm9326 (2022).
Terwilliger, T. C. et al. Accelerating crystal structure determination with iterative AlphaFold prediction. Acta Crystallogr. D 79, 234–244 (2023).
Blanc, M. et al. Designed ankyrin repeat proteins provide insights into the structure and function of CagI and are potent inhibitors of CagA translocation by the Helicobacter pylori type IV secretion system. PLoS Pathog. 19, e1011368 (2023).
Brookes, E., Rocco, M., Vachette, P. & Trewhella, J. AlphaFold-predicted protein structures and small-angle X-ray scattering: insights from an extended examination of selected data in the Small-Angle Scattering Biological Data Bank. J. Appl. Crystallogr. 56, 910–926 (2023).
Brookes, E. & Rocco, M. A database of calculated solution parameters for the AlphaFold predicted protein structures. Sci. Rep. 12, 7349 (2022).
Chinnam, N. B. et al. Combining small angle X-ray scattering (SAXS) with protein structure predictions to characterize conformations in solution. Methods Enzymol. 678, 351–376 (2023).
Da Vela, S. & Svergun, D. I. Methods, development and applications of small-angle X-ray scattering to characterize biological macromolecules in solution. Curr. Res. Struct. Biol. 2, 164–170 (2020).
Rambo, R. P. & Tainer, J. A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013).
Kryshtafovych, A. et al. Computational models in the service of X-ray and cryo-EM structure determination. Proteins 89, 1633–1646 (2021).
Chai, L. et al. AlphaFold protein structure database for sequence-independent molecular replacement. Crystals 11, 1227 (2021).
McCoy, A. J., Sammito, M. D. & Read, R. J. Implications of AlphaFold2 for crystallographic phasing by molecular replacement. Acta Crystallogr. D 78, 1–13 (2022).
Barbarin-Bocahu, I. & Graille, M. The X-ray crystallography phase problem solved thanks to AlphaFold and RoseTTAFold models: a case-study report. Acta Crystallogr. D 78, 517–531 (2022).
Abergel, C. Molecular replacement: tricks and treats. Acta Crystallogr. D 69, 2167–2173 (2013).
Chiliveri, S. C. et al. Experimental NOE, chemical shift, and proline isomerization data provide detailed insights into amelotin oligomerization. J. Am. Chem. Soc. 145, 18063–18074 (2023).
Abdollahi, H., Prestegard, J. H. & Valafar, H. Computational modeling multiple conformational states of proteins with residual dipolar coupling data. Curr. Opin. Struct. Biol. 82, 102655 (2023).
Sedinkin, S. L., Burns, D., Shukla, D., Potoyan, D. A. & Venditti, V. Solution structure ensembles of the open and closed forms of the ~130 kDa enzyme I via AlphaFold modeling, coarse grained simulations, and NMR. J. Am. Chem. Soc. 145, 13347–13356 (2023).
Li, E. H. et al. Blind assessment of monomeric AlphaFold2 protein structure models with experimental NMR data. J. Magn. Reson. 352, 107481 (2023).
Ma, P., Li, D.-W. & Brüschweiler, R. Predicting protein flexibility with AlphaFold. Proteins 91, 847–855 (2023).
Robertson, A. J., Courtney, J. M., Shen, Y., Ying, J. & Bax, A. Concordance of X-ray and AlphaFold2 models of SARS-CoV-2 main protease with residual dipolar couplings measured in solution. J. Am. Chem. Soc. 143, 19306–19310 (2021).
Lenard, A. J., Mulder, F. A. A. & Madl, T. Solvent paramagnetic relaxation enhancement as a versatile method for studying structure and dynamics of biomolecular systems. Prog. Nucl. Magn. Reson. Spectrosc. 132–133, 113–139 (2022).
Koehler Leman, J. & Künze, G. Recent advances in NMR protein structure prediction with Rosetta. Int. J. Mol. Sci. 24, 7835 (2023).
Zhu, W., Yang, D. T. & Gronenborn, A. M. Ligand-capped cobalt(II) multiplies the value of the double-histidine motif for PCS NMR studies. J. Am. Chem. Soc. 145, 4564–4569 (2023).
Klukowski, P., Riek, R. & Güntert, P. Time-optimized protein NMR assignment with an integrative deep learning approach using AlphaFold and chemical shift prediction. Sci. Adv. 9, eadi9323 (2023).
McShan, A. C. Utility of methyl side chain probes for solution NMR studies of large proteins. J. Magn. Reson. Open 14–15, 100087 (2023).
Ruschak, A. M. & Kay, L. E. Methyl groups as probes of supra-molecular structure, dynamics and function. J. Biomol. NMR 46, 75–87 (2009).
Pritišanac, I., Würz, J. M., Alderson, T. R. & Güntert, P. Automatic structure-based NMR methyl resonance assignment in large proteins. Nat. Commun. 10, 4922 (2019).
Clay, M. C., Saleh, T., Kamatham, S., Rossi, P. & Kalodimos, C. G. Progress toward automated methyl assignments for methyl-TROSY applications. Structure 30, 69–79 (2022).
Giri, N., Roy, R. S. & Cheng, J. Deep learning for reconstructing protein structures from cryo-EM density maps: recent advances and future directions. Curr. Opin. Struct. Biol. 79, 102536 (2023).
Hryc, C. F. & Baker, M. L. AlphaFold2 and cryoEM: revisiting cryoEM modeling in near-atomic resolution density maps. iScience 25, 104496 (2022).
Reggiano, G., Lugmayr, W., Farrell, D., Marlovits, T. C. & DiMaio, F. Residue-level error detection in cryo-electron microscopy models. Structure 31, 860–869 (2023).
Dai, X., Wu, L., Yoo, S. & Liu, Q. Integrating AlphaFold and deep learning for atomistic interpretation of cryo-EM maps. Brief. Bioinform. 24, bbad405 (2023).
Alshammari, M., He, J. & Wriggers, W. Appraisal of AlphaFold2-predicted models in cryo-EM map interpretation. Microsc. Microanal. 29, 977–978 (2023).
Lindorff-Larsen, K. & Kragelund, B. B. On the potential of machine learning to examine the relationship between sequence, structure, dynamics and function of intrinsically disordered proteins. J. Mol. Biol. 433, 167196 (2021).
Wei, G., Xi, W., Nussinov, R. & Ma, B. Protein ensembles: how does nature harness thermodynamic fluctuations for life? The diverse functional roles of conformational ensembles in the cell. Chem. Rev. 116, 6516–6551 (2016).
Sala, D., Hildebrand, P. W. & Meiler, J. Biasing AlphaFold2 to predict GPCRs and kinases with user-defined functional or structural properties. Front. Mol. Biosci. 10, 1121962 (2023).
Heo, L. & Feig, M. Multi-state modeling of G-protein coupled receptors at experimental accuracy. Proteins 90, 1873–1885 (2022).
Sala, D., Engelberger, F., Mchaourab, H. S. & Meiler, J. Modeling conformational states of proteins with AlphaFold. Curr. Opin. Struct. Biol. 81, 102645 (2023).
Stein, R. A. & Mchaourab, H. S. SPEACH_AF: sampling protein ensembles and conformational heterogeneity with AlphaFold2. PLoS Comput. Biol. 18, e1010483 (2022).
Wallner, B. AFsample: improving multimer prediction with AlphaFold using massive sampling. Bioinformatics 39, btad573 (2023).
Johansson-Åkhe, I. & Wallner, B. Improving peptide–protein docking with AlphaFold-Multimer using forced sampling. Front. Bioinform. 2, 959160 (2022).
Ramelot, T. A., Tejero, R. & Montelione, G. T. Representing structures of the multiple conformational states of proteins. Curr. Opin. Struct. Biol. 83, 102703 (2023).
Townshend, R. J. L. et al. Geometric deep learning of RNA structure. Science 373, 1047–1051 (2021).
Bojar, D. & Lisacek, F. Glycoinformatics in the artificial intelligence era. Chem. Rev. 122, 15971–15988 (2022).
Acknowledgements
A.C.M. acknowledges start-up funds from the Georgia Institute of Technology. V.A. acknowledges support from the National Science Foundation (CHE-2238650) and the National Institutes of Health (R35GM142882).
Author information
Authors and Affiliations
Contributions
A.C.M. and V.A. conceived, wrote and edited the manuscript. A.C.M. generated figures and analyzed models.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Paul Adams and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Agarwal, V., McShan, A.C. The power and pitfalls of AlphaFold2 for structure prediction beyond rigid globular proteins. Nat Chem Biol 20, 950–959 (2024). https://doi.org/10.1038/s41589-024-01638-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-024-01638-w
This article is cited by
-
Large-scale predictions of alternative protein conformations by AlphaFold2-based sequence association
Nature Communications (2025)
-
Structure and mechanism of human vesicular polyamine transporter
Nature Communications (2025)
-
Integrating chemical artificial intelligence and cognitive computing for predictive analysis of biological pathways: a case for intrinsically disordered proteins
Biophysical Reviews (2025)
-
Structural Insights into Cold-Active Lipase from Glaciozyma antarctica PI12: Alphafold2 Prediction and Molecular Dynamics Simulation
Journal of Molecular Evolution (2024)