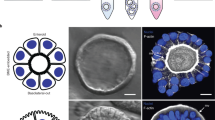
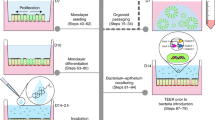
Abstract
Adult-stem-cell-derived organoids model human epithelial tissues ex vivo, which enables the study of host–microbe interactions with great experimental control. This protocol comprises methods to coculture organoids with microbes, particularly focusing on human small intestinal and colon organoids exposed to individual bacterial species. Microinjection into the lumen and periphery of 3D organoids is discussed, as well as exposure of organoids to microbes in a 2D layer. We provide detailed protocols for characterizing the coculture with regard to bacterial and organoid cell viability and growth kinetics. Spatial relationships can be studied by fluorescence live microscopy, as well as scanning electron microscopy. Finally, we discuss considerations for assessing the impact of bacteria on gene expression and mutations through RNA and DNA sequencing. This protocol requires equipment for standard mammalian tissue culture, or bacterial or viral culture, as well as a microinjection device.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All previously unpublished data are included in the figures. Raw image files are available from the corresponding author upon request.
References
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Dutta, D., Heo, I. & Clevers, H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 23, 393–410 (2017).
Min, S., Kim, S. & Cho, S.-W. Gastrointestinal tract modeling using organoids engineered with cellular and microbiota niches. Exp. Mol. Med. 52, 227–237 (2020).
Leslie, J. L. & Young, V. B. A whole new ball game: stem cell-derived epithelia in the study of host-microbe interactions. Anaerobe 37, 25–28 (2016).
Heo, I. et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 3, 814–823 (2018).
Bartfeld, S. et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148, 126–136.e6 (2015).
Ettayebi, K. et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393 (2016).
Lamers, M. M. et al. SARS-CoV-2 productively infects human gut enterocytes. Science https://doi.org/10.1126/science.abc1669 (2020).
Zhou, J. et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 26, 1077–1083 (2020).
Pleguezuelos-Manzano, C. et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580, 269–273 (2020).
Andersson-Rolf, A., Fink, J., Mustata, R. C. & Koo, B.-K. A video protocol of retroviral infection in primary intestinal organoid culture. J. Vis. Exp. https://doi.org/10.3791/51765 (2014).
Holthaus, D., Delgado-Betancourt, E., Aebischer, T., Seeber, F. & Klotz, C. Harmonization of protocols for multi-species organoid platforms to study the intestinal biology of Toxoplasma gondii and other protozoan infections. Front. Cell. Infect. Microbiol. 10, 610368 (2021).
Dutta, D., Heo, I. & O’Connor, R. Studying Cryptosporidium infection in 3D tissue-derived human organoid culture systems by microinjection. J. Vis. Exp. https://doi.org/10.3791/59610 (2019).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011).
Fakhiri, J. et al. Novel chimeric gene therapy vectors based on adeno-associated virus and four different mammalian bocaviruses. Mol. Ther. Methods Clin. Dev. 12, 202–222 (2019).
Pleguezuelos‐Manzano, C. et al. Establishment and culture of human intestinal organoids derived from adult stem cells. Curr. Protoc. Immunol. 130, (2020).
Beumer, J. et al. High-resolution mRNA and secretome atlas of human enteroendocrine cells. Cell 181, 1291–1306.e19 (2020).
Boonekamp, K. E., Dayton, T. L. & Clevers, H. Intestinal organoids as tools for enriching and studying specific and rare cell types: advances and future directions. J. Mol. Cell Biol. https://doi.org/10.1093/jmcb/mjaa034 (2020).
Bar-Ephraim, Y. E., Kretzschmar, K. & Clevers, H. Organoids in immunological research. Nat. Rev. Immunol. 20, 279–293 (2020).
Dijkstra, K. K. et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell 174, 1586–1598.e12 (2018).
Greicius, G. et al. PDGFRα+ pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc. Natl Acad. Sci. USA 115, E3173–E3181 (2018).
Noel, G. et al. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 7, 45270 (2017).
Round, J. L. & Mazmanian, S. K. The gut microbiome shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 (2009).
Spence, J. R. et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 (2011).
McCauley, H. A. & Wells, J. M. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 144, 958–962 (2017).
Forbester, J. L. et al. Interaction of Salmonella enterica serovar Typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect. Immun. 83, 2926–2934 (2015).
Holokai, L. et al. Increased programmed death ligand-1 is an early epithelial cell response to Helicobacter pylori infection. PLoS Pathog. 15, e1007468 (2019).
Finkbeiner, S. R. et al. Stem cell–derived human intestinal organoids as an infection model for rotaviruses. mBio 3, e00159-12 (2012).
Nikolaev, M. et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 585, 574–578 (2020).
Jalili-Firoozinezhad, S. et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 3, 520–531 (2019).
Steinway, S. N., Saleh, J., Koo, B.-K., Delacour, D. & Kim, D.-H. Human microphysiological models of intestinal tissue and gut microbiome. Front. Bioeng. Biotechnol. 8, 725 (2020).
Hinman, S. S., Wang, Y. & Allbritton, N. L. Photopatterned membranes and chemical gradients enable scalable phenotypic organization of primary human colon epithelial models. Anal. Chem. 91, 15240–15247 (2019).
Avraham, R. et al. A highly multiplexed and sensitive RNA-seq protocol for simultaneous analysis of host and pathogen transcriptomes. Nat. Protoc. 11, 1477–1491 (2016).
Marsh, J. W., Humphrys, M. S. & Myers, G. S. A. A laboratory methodology for dual RNA-sequencing of bacteria and their host cells in vitro. Front. Microbiol. 8, 1830 (2017).
Westermann, A. J. et al. Dual RNA-seq unveils noncoding RNA functions in host–pathogen interactions. Nature 529, 496–501 (2016).
Sezonov, G., Joseleau-Petit, D. & D’Ari, R. Escherichia coli physiology in Luria–Bertani broth. J. Bacteriol. 189, 8746–8749 (2007).
Sim, K. et al. Improved detection of bifidobacteria with optimised 16S rRNA-gene based pyrosequencing. PLoS ONE 7, e32543 (2012).
Blokzijl, F. et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016).
Acknowledgements
We thank E. Allen-Vercoe and A. Robinson for provision of bacterial strains and discussions on bacterial culturing conditions. The development of the methods was supported by CRUK grant OPTIMISTICC (C10674/A27140) (J.P., C.P.-M. and H.C.), the Gravitation projects CancerGenomiCs.nl, and the Netherlands Organ-on-Chip Initiative (024.003.001) from the Netherlands Organisation for Scientific Research (NWO) funded by the Ministry of Education, Culture and Science of the government of the Netherlands (J.P., C.P.-M. and H.C.), the Oncode Institute (partly financed by the Dutch Cancer Society), the European Research Council under ERC Advanced Grant Agreement no. 67013 (J.P., D.D., I.H. and H.C.) and NETRF/Petersen Accelerator (J.B.).
Author information
Authors and Affiliations
Contributions
All authors contributed to the development of the organoid co-culture methods described in this protocol. J.P., C.P.M. and H.C. wrote the manuscript with input and corrections from all authors. A.M.S., C.P.M. and J.P. prepared figures.
Corresponding author
Ethics declarations
Competing interests
H.C. is inventor on multiple patents held by the Dutch Royal Netherlands Academy of Arts and Sciences that cover organoid technology: PCT/NL2008/050543, WO2009/022907; PCT/NL2010/000017, WO2010/090513; PCT/IB2011/002167, WO2012/014076; PCT/IB2012/052950, WO2012/168930; PCT/EP2015/060815, WO2015/173425; PCT/EP2015/077990, WO2016/083613; PCT/EP2015/077988, WO2016/083612; PCT/EP2017/054797,WO2017/149025; PCT/EP2017/065101, WO2017/220586; PCT/EP2018/086716; and GB1819224.5. H.C.’s full disclosure is given at https://www.uu.nl/staff/JCClevers/.
Additional information
Peer review information Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Pleguezuelos-Manzano, C. et al. Nature 580, 269–273 (2020): https://doi.org/10.1038/s41586-020-2080-8
Heo, I. et al. Nat. Microbiol. 3, 814–823 (2018): https://doi.org/10.1038/s41564-018-0177-8
Lamers, M. M. et al. Science 369, 50–54 (2020): https://doi.org/10.1126/science.abc1669
Supplementary information
Rights and permissions
About this article
Cite this article
Puschhof, J., Pleguezuelos-Manzano, C., Martinez-Silgado, A. et al. Intestinal organoid cocultures with microbes. Nat Protoc 16, 4633–4649 (2021). https://doi.org/10.1038/s41596-021-00589-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00589-z
This article is cited by
-
Decoding host-microbe interactions with engineered human organoids
The EMBO Journal (2025)
-
Dual RNA sequencing of a co-culture model of Pseudomonas aeruginosa and human 2D upper airway organoids
Scientific Reports (2025)
-
Imaging 3D cell cultures with optical microscopy
Nature Methods (2025)
-
Adapting systems biology to address the complexity of human disease in the single-cell era
Nature Reviews Genetics (2025)
-
Model systems to study tumor-microbiome interactions in early-onset colorectal cancer
EMBO Molecular Medicine (2025)