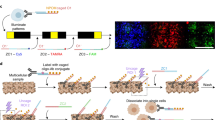
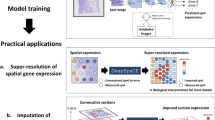
Abstract
Spatial transcriptomics technologies aim to advance gene expression studies by profiling the entire transcriptome with intact spatial information from a single histological slide. However, the application of spatial transcriptomics is limited by low resolution, limited transcript coverage, complex procedures, poor scalability and high costs of initial setup and/or individual experiments. Seq-Scope repurposes the Illumina sequencing platform for high-resolution, high-content spatial transcriptome analysis, overcoming these limitations. It offers submicrometer resolution, high capture efficiency, rapid turnaround time and precise annotation of histopathology at a much lower cost than commercial alternatives. This protocol details the implementation of Seq-Scope with an Illumina NovaSeq 6000 sequencing flow cell, allowing the profiling of multiple tissue sections in an area of 7 mm × 7 mm or larger. We describe the preparation of a fresh-frozen tissue section for both histological imaging and sequencing library preparation and provide a streamlined computational pipeline with comprehensive instructions to integrate histological and transcriptomic data for high-resolution spatial analysis. This includes the use of conventional software tools for single-cell and spatial analysis, as well as our recently developed segmentation-free method for analyzing spatial data at submicrometer resolution. Aside from array production and sequencing, which can be done in batches, tissue processing, library preparation and running the computational pipeline can be completed within 3 days by researchers with experience in molecular biology, histology and basic Unix skills. Given its adaptability across various biological tissues, Seq-Scope establishes itself as an invaluable tool for researchers in molecular biology and histology.
Key points
-
The protocol repurposes an Illumina NovaSeq 6000 flow cell for spatial transcriptomics, generating high-resolution datasets and integrating a streamlined data-analysis pipeline.
-
Leveraging commonly available Illumina equipment, the protocol offers researchers ultra-high, submicrometer resolution in spatial transcriptomics analysis with a comprehensive analytical pipeline, whole-transcriptome coverage, rapid turnaround, cost efficiency and versatility.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
























Similar content being viewed by others
Data availability
All source data described in this protocol are available online via public repositories, such as Zenodo and Deep Blue Data: minimal test run dataset (https://doi.org/10.5281/zenodo.10835761), shallow-sequenced liver dataset (https://doi.org/10.5281/zenodo.10840696), deep-sequenced liver dataset (https://doi.org/10.7302/tw62-4f97) and exemplary downstream analysis input (https://doi.org/10.5281/zenodo.10841777).
Code availability
All source code described in this protocol is available online via GitHub: NovaScope pipeline, v1.0.0 (https://github.com/seqscope/novascope) and NEDA, v1.0.0 (https://github.com/seqscope/NovaScope-exemplary-downstream-analysis).
References
Stahl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Bergenstrahle, J., Larsson, L. & Lundeberg, J. Seamless integration of image and molecular analysis for spatial transcriptomics workflows. BMC Genomics 21, 482 (2020).
Stickels, R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 39, 313–319 (2021).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665–1681.e18 (2020).
Sergio Marco, S. et al. Optimizing xenium in situ data utility by quality assessment and best practice analysis workflows. Preprint at https://www.biorxiv.org/content/10.1101/2023.02.13.528102v1 (2023).
He, S. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 40, 1794–1806 (2022).
Zhang, M. et al. Molecularly defined and spatially resolved cell atlas of the whole mouse brain. Nature 624, 343–354 (2023).
Slovin, S. et al. Single-cell RNA sequencing analysis: a step-by-step overview. Methods Mol. Biol. 2284, 343–365 (2021).
Cho, C.-S. et al. Microscopic examination of spatial transcriptome using Seq-Scope. Cell 184, 3559–3572.e22 (2021).
10x Genomics. Visium HD: whole transcriptome spatial discovery at the resolution you need. https://www.10xgenomics.com/library/8012d2 (accessed 25 February 2024).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792.e21 (2022).
Fu, X. et al. Polony gels enable amplifiable DNA stamping and spatial transcriptomics of chronic pain. Cell 185, 4621–4633.e17 (2022).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
BD_Biosciences BD Rhapsody™ system—mRNA whole transcriptome analysis (WTA) library preparation protocol. https://scomix.bd.com/hc/article_attachments/13726971063565 (2022).
Salmen, F. et al. Barcoded solid-phase RNA capture for spatial transcriptomics profiling in mammalian tissue sections. Nat. Protoc. 13, 2501–2534 (2018).
10x Genomics. Visium spatial tissue optimization reagents kits user guide. https://www.10xgenomics.com/support/spatial-gene-expression-fresh-frozen/documentation/steps/tissue-optimization/visium-spatial-tissue-optimization-reagents-kits-user-guide (2022).
10x Genomics. Visium spatial gene expression reagent kits user guide. https://www.10xgenomics.com/support/spatial-gene-expression-fresh-frozen/documentation/steps/library-construction/visium-spatial-gene-expression-reagent-kits-user-guide (2023).
Kaminow, B., Yunosov, D. & Dobin, A. STARsolo: accurate, fast and versatile mapping/quantification of single-cell and single-nucleus RNA-seq data. Preprint at https://www.biorxiv.org/content/10.1101/2021.05.05.442755v1 (2021).
Xi, J., Lee, J. H., Kang, H. M. & Jun, G. STtools: a comprehensive software pipeline for ultra-high-resolution spatial transcriptomics data. Bioinform. Adv. 2, vbac061 (2022).
Do, T. H. et al. TREM2 macrophages induced by human lipids drive inflammation in acne lesions. Sci. Immunol. 7, eabo2787 (2022).
Ma, F. et al. Single cell and spatial sequencing define processes by which keratinocytes and fibroblasts amplify inflammatory responses in psoriasis. Nat. Commun. 14, 3455 (2023).
Hsu, J. et al. High-resolution spatial transcriptomic atlas of mouse soleus muscle: unveiling single cell and subcellular heterogeneity in health and denervation. Preprint at https://www.biorxiv.org/content/10.1101/2024.02.26.582103v1 (2024).
Poovathingal, S. et al. Nova-ST: Nano-patterned ultra-dense platform for spatial transcriptomics. Cell Rep. Methods 4, 100831 (2024).
Schott, M. et al. Open-ST: high-resolution spatial transcriptomics in 3D. Cell 187, 3953–3972.e26 (2024).
Bahar Halpern, K. et al. Nuclear retention of mRNA in mammalian tissues. Cell Rep. 13, 2653–2662 (2015).
Gurkar, A. U. et al. Spatial mapping of cellular senescence: emerging challenges and opportunities. Nat. Aging 3, 776–790 (2023).
Cao, J. et al. Decoder-seq enhances mRNA capture efficiency in spatial RNA sequencing. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-02086-y (2024).
Bai, Z. et al. Spatially exploring RNA biology in archival formalin-fixed paraffin-embedded tissues. Cell https://doi.org/10.1016/j.cell.2024.09.001 (2024).
Wulf, M. G. et al. Non-templated addition and template switching by Moloney murine leukemia virus (MMLV)-based reverse transcriptases co-occur and compete with each other. J. Biol. Chem. 294, 18220–18231 (2019).
Hughes, T. K. et al. Second-strand synthesis-based massively parallel scRNA-seq reveals cellular states and molecular features of human inflammatory skin pathologies. Immunity 53, 878–894.e7 (2020).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Si, Y. et al. FICTURE: scalable segmentation-free analysis of submicron-resolution spatial transcriptomics. Nat. Methods 21, 1843–1854 (2024).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Pierre, S. & Luc, M. V. Determining watersheds in digital pictures via flooding simulations. Proc. SPIE Int. Soc. Opt. Eng. 1360, 240–250 (1990).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Pachitariu, M. & Stringer, C. Cellpose 2.0: how to train your own model. Nat. Methods 19, 1634–1641 (2022).
Dries, R. et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 22, 78 (2021).
Palla, G. et al. Squidpy: a scalable framework for spatial omics analysis. Nat. Methods 19, 171–178 (2022).
Zhang, D. et al. Spatial epigenome-transcriptome co-profiling of mammalian tissues. Nature 616, 113–122 (2023).
Llorens-Bobadilla, E. et al. Solid-phase capture and profiling of open chromatin by spatial ATAC. Nat. Biotechnol. 41, 1085–1088 (2023).
Deng, Y. et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature 609, 375–383 (2022).
Vickovic, S. et al. SM-Omics is an automated platform for high-throughput spatial multi-omics. Nat. Commun. 13, 795 (2022).
Zhao, T. et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature 601, 85–91 (2022).
Engblom, C. et al. Spatial transcriptomics of B cell and T cell receptors reveals lymphocyte clonal dynamics. Science 382, eadf8486 (2023).
Benotmane, J. K. et al. High-sensitive spatially resolved T cell receptor sequencing with SPTCR-seq. Nat. Commun. 14, 7432 (2023).
Hudson, W. H. & Sudmeier, L. J. Localization of T cell clonotypes using the Visium spatial transcriptomics platform. STAR Protoc. 3, 101391 (2022).
Liu, Y., Enninful, A., Deng, Y. & Fan, R. Spatial transcriptome sequencing of FFPE tissues at cellular level. Preprint at https://www.biorxiv.org/content/10.1101/2020.10.13.338475v2.full (2020).
Gracia Villacampa, E. et al. Genome-wide spatial expression profiling in formalin-fixed tissues. Cell Genom. 1, 100065 (2021).
Heiser, C. N. et al. Molecular cartography uncovers evolutionary and microenvironmental dynamics in sporadic colorectal tumors. Cell 186, 5620–5637.e16 (2023).
Illumina sequencing library QC on the MiSeq system. https://www.illumina.com/content/dam/illumina-marketing/documents/products/appnotes/appnote_miseq_libqc.pdf (2014).
Illumina bcl2fastq2 conversion software v2.20 software guide. https://support.illumina.com/content/dam/illumina-support/documents/documentation/software_documentation/bcl2fastq/bcl2fastq2-v2-20-software-guide-15051736-03.pdf (2019).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Lako, A. & Rodig, S. HTAPP_hematoxylin and eosin (H&E) staining protocol of OCT frozen tissue. https://doi.org/10.17504/protocols.io.8pphvmn (2019).
Ma, Z. Y. et al. Comparison of staining quality between rapid and routine hematoxylin and eosin staining of frozen breast tissue sections: an observational study. J. Int. Med. Res. 52, 3000605241259682 (2024).
Molder, F. et al. Sustainable data analysis with Snakemake. F1000Res 10, 33 (2021).
Acknowledgements
The work was supported by the Taubman Institute Innovation Project (to H.M.K. and J.H.L.), the NIH (T32AG000114 to Y.K. and C.-S.C., K01AG061236 to M.K., R01DK118631 to G.J., R01AG079163 to M.K. and J.H.L., R01HG011031 and HHSN268201800002I to H.M.K. and R01DK133448 and UH3CA268091 to J.H.L.), Technology Transfer Talent Network (T3N) Postdoctoral Fellowship (to Y.K.) and a Gleen Foundation Core grant (to J.H.L.).
Author information
Authors and Affiliations
Contributions
Y.K., C.-S.C., A.P., M.S., J.-E.H., M.K. and J.H.L. developed the experimental part of the protocol. W.C., Y.H., Y.S., J.X., A.A., C.L., G.J. and H.M.K. developed the computational part of the protocol. E.P., O.I.K., T.W., H.M.K. and J.H.L. developed the sequencing part of the protocol. Y.K., W.C., C.-S.C., H.M.K. and J.H.L. prepared the first draft. All authors revised, reviewed and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
H.M.K. owns stock in Regeneron Pharmaceuticals. J.H.L. is an inventor on a patent and pending patent applications related to Seq-Scope.
Peer review
Peer review information
Nature Protocols thanks Moritz Gerstung and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Cho, C.-S. et al. Cell 184, 3559–3572.e22 (2021): https://doi.org/10.1016/j.cell.2021.05.010
Xi, J. et al. Bioinform. Adv. 2, vbac061 (2022): https://doi.org/10.1093/bioadv/vbac061
Si, Y. et al. Nat. Methods 21, 1843–1854 (2024): https://doi.org/10.1038/s41592-024-02415-2
Do, T. H. et al. Sci. Immunol. 7, eabo2787 (2022): https://doi.org/10.1126/sciimmunol.abo2787
Supplementary information
Supplementary Data 1
3D model (STL) file for the custom frame adapter described in the protocol. The STL file can be used to fabricate the adapter in most 3D printing service centers. We printed the adapter on a Stratasys J850 by using the default white material.
Supplementary Data 2
Sketch drawing and specification of the custom silicone isolator (Grace Bio-Labs, cat. no. JTR25-A-1.0, RD501346). Information in this PDF is sufficient to reproduce the part with the same specifications applied by Grace Bio-Labs.
Supplementary Data 3
README file providing details on running the code and software applied in the protocol
Supplementary Video 1
NovaSeq 6000 S4 flow cell disassembly. The scalpel is used to separate the flow cell into its three main components. This video demonstrates the entire procedure of the flow cell disassembly. During the demonstration, viewers may notice that a small piece was broken off the top layer of the flow cell. This layer is thin and, therefore, prone to breakage. However, as long as the broken piece is outside the imaging area described in Fig. 3d (B02–B10 and T02–T10), it does not interfere with subsequent procedures. Furthermore, even if the breakage damages some imaging areas, other intact areas can still be effectively used without any issues.
Supplementary Video 2
Seq-Scope chip dicing procedure. The chip was diced from the disassembled top and bottom layers of a NovaSeq 6000 S4 flow cell by using a precision glass cutter.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kim, Y., Cheng, W., Cho, CS. et al. Seq-Scope: repurposing Illumina sequencing flow cells for high-resolution spatial transcriptomics. Nat Protoc 20, 643–689 (2025). https://doi.org/10.1038/s41596-024-01065-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-024-01065-0
This article is cited by
-
Spatial omics technology potentially promotes the progress of tumor immunotherapy
British Journal of Cancer (2025)