Figure 1
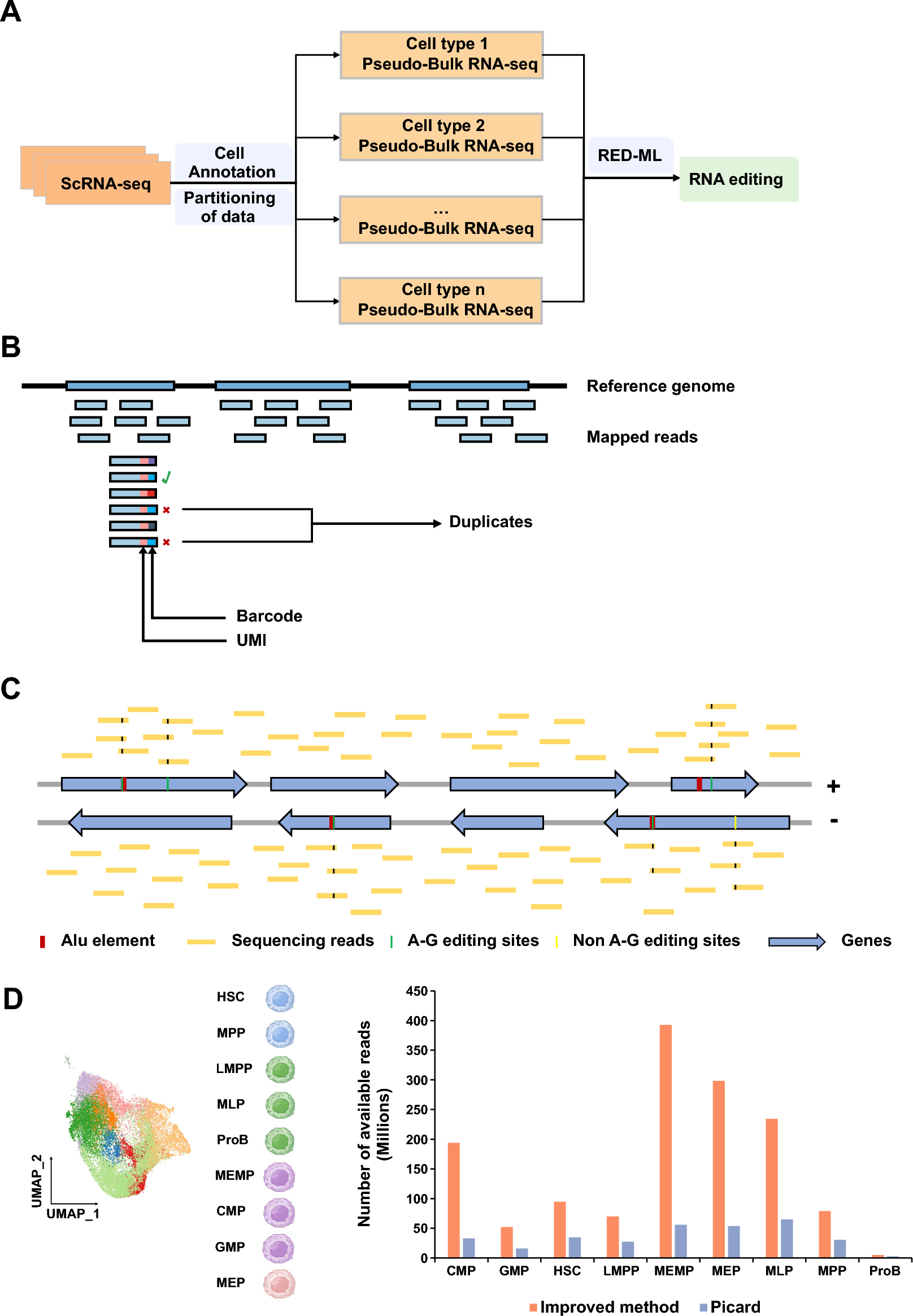
A schematic diagram showing the computational method developed in this study. (A) A workflow showing the strategies to identify RNA editing events using scRNA-seq. The cell type annotation information was used to combine the mapped reads of the same cell type in scRNA-seq to obtain pseudo-Bulk RNA-seq for each cell type. (B) The novel threshold used to mark duplicates. Only if the aligned reads with the same alignment position, UMI and barcode are defined as duplicates (see details in “Methods” section). (C) The reads aligned to the reference genome are divided into reads aligned to the forward strand and reverse strand to distinguish RNA editing sites occurring on the forward/reverse strand genes. The edited sites located in ALU element and with the A-G variation are more likely to be identified as RNA editing sites. (D) The scRNA-seq data of hematopoietic stem cells (HSCs), multipotential progenitor cells (MPPs), lymphoid lineage multipotential progenitor cells (LMPPs), multipotential lymphoid lineage progenitor cells (MLPs), megakaryocyte erythroid mast progenitor cells (MEMPs), common myeloid progenitor cells (CMPs), granulocyte monocyte progenitors (GMPs), megakaryocyte erythroid progenitors (MEPs), and B-cell progenitors (ProBs) were used to evaluate improved method and Picard. Bar plot showing the number of available reads using improved method and Picard.
