Abstract
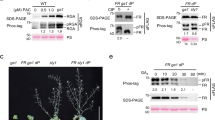
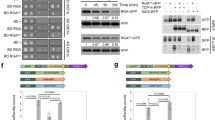
Light and gibberellins (GAs) mediate many essential and partially overlapping plant developmental processes. DELLA proteins are GA-signalling repressors that block GA-induced development1. GA induces degradation of DELLA proteins via the ubiquitin/proteasome pathway2, but light promotes accumulation of DELLA proteins by reducing GA levels3. It was proposed that DELLA proteins restrain plant growth largely through their effect on gene expression4,5. However, the precise mechanism of their function in coordinating GA signalling and gene expression remains unknown. Here we characterize a nuclear protein interaction cascade mediating transduction of GA signals to the activity regulation of a light-responsive transcription factor. In the absence of GA, nuclear-localized DELLA proteins accumulate to higher levels, interact with phytochrome-interacting factor 3 (PIF3, a bHLH-type transcription factor) and prevent PIF3 from binding to its target gene promoters and regulating gene expression, and therefore abrogate PIF3-mediated light control of hypocotyl elongation. In the presence of GA, GID1 proteins (GA receptors) elevate their direct interaction with DELLA proteins in the nucleus, trigger DELLA protein’s ubiquitination and proteasome-mediated degradation, and thus release PIF3 from the negative effect of DELLA proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
199,00 € per year
only 3,90 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fleet, C. M. & Sun, T. P. A. DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8, 77–85 (2005)
Itoh, H., Matsuoka, M. & Steber, C. M. A role for the ubiquitin–26S-proteasome pathway in gibberellin signaling. Trends Plant Sci. 8, 492–497 (2003)
Achard, P. et al. DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143, 1163–1172 (2007)
Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 218, 683–692 (2004)
Cao, D., Cheng, H., Wu, W., Soo, H. M. & Peng, J. Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis . Plant Physiol. 142, 509–525 (2006)
Alabadi, D., Gil, J., Blazquez, M. A. & Garcia-Martinez, J. L. Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 134, 1050–1057 (2004)
Dill, A., Thomas, S. G., Hu, J., Steber, C. M. & Sun, T. P. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16, 1392–1405 (2004)
Fu, X. et al. The Arabidopsis mutant sleepy1gar2–1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16, 1406–1418 (2004)
Silverstone, A. L. et al. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis . Plant Cell 13, 1555–1566 (2001)
Itoh, H., Ueguchi-Tanaka, M., Sato, U., Ashikari, M. & Matsuoka, M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70 (2002)
Kim, J. et al. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15, 2399–2407 (2003)
Martinez-Garcia, J. F., Huq, E. & Quail, P. H. Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863 (2000)
Ni, M., Tepperman, J. M. & Quail, P. H. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784 (1999)
Zhu, Y., Tepperman, J. M., Fairchild, C. D. & Quail, P. H. Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix–loop–helix factor PIF3 in a reaction requiring the PAS ___domain of PIF3. Proc. Natl Acad. Sci. USA 97, 13419–13424 (2000)
Bauer, D. et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis . Plant Cell 16, 1433–1445 (2004)
Park, E. et al. Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45, 968–975 (2004)
Al-Sady, B., Ni, W., Kircher, S., Schafer, E. & Quail, P. H. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446 (2006)
Lee, J. et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749 (2007)
Jiao, Y., Ma, L., Strickland, E. & Deng, X. W. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis . Plant Cell 17, 3239–3256 (2005)
Monte, E. et al. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl Acad. Sci. USA 101, 16091–16098 (2004)
Ogawa, M. et al. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15, 1591–1604 (2003)
Ueguchi-Tanaka, M. et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698 (2005)
Nakajima, M. et al. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46, 880–889 (2006)
Griffiths, J. et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis . Plant Cell 18, 3399–3414 (2007)
Willige, B. C. et al. The DELLA ___domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis . Plant Cell 19, 1209–1220 (2007)
Toledo-Ortiz, G., Huq, E. & Quail, P. H. The Arabidopsis basic/helix–loop–helix transcription factor family. Plant Cell 15, 1749–1770 (2003)
de Lucas, M. et al. A molecular framework for light and gibberellin control of cell elongation. Nature doi: 10.1038/nature06520
Oh, E. et al. PIL5, a phytochrome-interacting basic helix–loop–helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana . Plant Cell 16, 3045–3058 (2004)
Penfield, S. et al. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15, 1998–2006 (2005)
Feng, S. et al. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 16, 1870–1882 (2004)
Rubio, V. et al. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41, 767–778 (2005)
Feng, S. et al. The COP9 signalosome interacts physically with SCFCOI1 and modulates jasmonate responses. Plant Cell 15, 1083–1094 (2003)
Acknowledgements
We thank J. Lee, K. He and I. Lee for providing unpublished results of their ChIP-microarray studies; J. A. Sullivan for MYC- and HA-tag vectors; F. Nagy for the YFP-tag vector; X. P. Wang for the Flag-tag vector; P.H. Quail for the pPIF3-RSETb plasmid; G. Choi for pif3-1 and 35S-PIF3–His–MYC seeds; T. P. Sun for rga-24 and rgl2-13 seeds; N. P. Harberd for gai-t6 seeds; S. P. Dinesh-Kumar for tobacco seeds; and F. Parcy for providing the BiFC vector system. This work was supported by a grant from the National Institutes of Health to X.W.D., by a grant from the Ministry of Science and Technology of China to National Institute of Biological Sciences at Beijing, by a 985 program fund from Peking University and the Ministry of Education of China to the Peking-Yale Joint Center laboratory, and by a grant from Program for New Century Excellent Talents in Peking University to L.-M.F. C.M. is a recipient of a postdoctoral fellowship from the Spanish Ministerio de Educacion y Ciencia. J.M.I.-P. is a recipient of a predoctoral fellowship from the Spanish Ministerio de Educacion y Ciencia. E.S. and S.K. were supported by grants from the Deutsche Forschungsgesellschaft.
Author Contributions X.W.D. conceived the project, and S.F. and X.W.D. together designed the experiments. L.-M.F. designed some of the experiments. S.F. and G.G. performed chromatin immunoprecipitation. C.M. and G.G. analysed the gid1 mutants and performed RT–PCR. Y.W. made the anti-RGA antibody. Y.W., L.C., F.W. and L.Y. performed the yeast two-hybrid analyses. J.Z. and F.W. performed in vitro pull-down assays. C.M. and J.M.I.-P. performed the BiFC experiments. S.K. and E.S. performed phyB and PIF3 subcellular localization studies. X.F. provided the rga-24 gai-t6 double and della pentuple mutants. S.F. performed all other experiments. S.F. and X.W.D. wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Notes, Supplementary Figures 1-7 with Legends and Supplementary Tables 1-2. (PDF 2930 kb)
Rights and permissions
About this article
Cite this article
Feng, S., Martinez, C., Gusmaroli, G. et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479 (2008). https://doi.org/10.1038/nature06448
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06448
This article is cited by
-
Petunia PHYTOCHROME INTERACTING FACTOR 4/5 transcriptionally activates key regulators of floral scent
Plant Molecular Biology (2024)
-
OsCSN2 orchestrates Oryza sativa L. growth and development through modulation of the GA and BR pathways
Functional & Integrative Genomics (2024)
-
Uncovering the involvement of DoDELLA1-interacting proteins in development by characterizing the DoDELLA gene family in Dendrobium officinale
BMC Plant Biology (2023)
-
The Arabidopsis endosperm is a temperature-sensing tissue that implements seed thermoinhibition through phyB
Nature Communications (2023)
-
The master growth regulator DELLA binding to histone H2A is essential for DELLA-mediated global transcription regulation
Nature Plants (2023)