Abstract
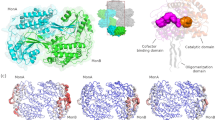
Aldehyde reductase, a member of the aldo-keto reductase superfamily, catalyzes the NADPH-dependent reduction of a variety of aldehydes to their corresponding alcohols. The structure of porcine aldehyde reductase–NADPH binary complex has been determined by X-ray diffraction methods and refined to a crystallographic R-factor of 0.20 at 2.4 Å resolution. The tertiary structure of aldehyde reductase is similar to that of aldose reductase and consists of an α/β-barrel with the active site located at the carboxy terminus of the strands of the barrel. Unlike aldose reductase, the Nε2 of the imidazole ring of His 113 in aldehyde reductase interacts, through a hydrogen bond, with the amide group of the nicotinamide ring of NADPH.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Flynn, T.G. Aldehyde reductases: monomeric NADPH-dependent oxidoreductases with multifunctional potential. Biochem. Pharmac. 31, 2705–2712 (1982).
Wermuth, B. in Enzymology and molecular biology of carbonyl metabolism (Eds Flynn, T G. & Weiner, H.) 209–230 (Alan, R. Liss, New York; 1985).
Bohren, K.M., Bullock, B., Wermuth, B. & Gabbay, K.H. The aldo-keto reductase superfamily. J. biol. Chem. 264, 9547–9551 (1989).
Kinoshita, J.H. & Nishimura, C. The involvement of aldose reductase in diabetic complications. Diabetes metab. Rev. 4, 323–337 (1988).
Srivastava, S.K., Petrash, J.M., Sadana, I.J., Ansari, N.H. & Partridge, C.A. Susceptibility of aldehyde and aldose reductases of human tissues to aldose redutase inhibitors. Curr. Eye Res. 83, 407–410 (1982).
Spielberg, S.P., Shear, N.H., Cannon, M., Huston, N.J. & Gunderson, K. In-vitro assessment of a hypersensitivity syndrome associated with sorbinil. Anal. intern. Med. 114, 720–724 (1991).
Sarges, R. & Oates, P. Aldose reductase inhibitors: recent developments. J. Prog. Drug Res. 40, 99–161 (1993).
Rondeau, J.-M. et al. Novel NADPH binding ___domain revealed by the crystal structure of aldose reductase. Nature 355, 469–472 (1992).
Wilson, D.K., Bohren, K.M., Gabbay, K.H. & Quiocho, F.A. An unlikely sugar substrate site in the 1.65 Å structure of human aldose reductase holoenzyme implicated in diabetic complications. Science 257, 81–84 (1992).
Grimshaw, C.E., Shahbaz, M. & Putney, C.G. Mechanistic basis for non-linear kinetics of aldehyde reduction catalyzed by aldose reductase. Biochemistry 29, 9947–9955 (1990).
Kubiseski, T.J., Hyndman, D.J., Morjana, N.A. & Flynn, T.G. Studies on pig muscle aldose reductase. Kinetic mechanism and evidence for a slow conformational change upon coenzyme binding. J. biol. Chem. 267, 6510–6517 (1992).
Kubiseski, T.J., Green, N.C., Borhani, D.W. & Flynn, T.G. Studies on pig aldose reductase. Identification of an essential arginine in the primary and tertiary structure of the enzyme. J. biol. Chem. 2183–2188 (1994).
Harrison, D.H., Bohren, K.M., Ringe, D., Petsko, G.A. & Gabbay, K.H. An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cocodylate, and glucose 6-phosphate. Biochemistry 33, 2011–2020 (1994).
El-Kabbani, O. et al Structures of human and porcine aldehyde reductase: an enzyme implicated in diabetic complications. Acta Crystallogr. D50, 859–868 (1994).
Farber, G.K. & Petsko, G.A. The evolution of α/β barrel enzymes. Trends Biochem. Sci. 15, 228–234 (1990).
Wermuth, B. Enzymology and Molecular Biology of Carbonyl Metabolism (Eds Weiner, H. & Wermuth, B.) 261–274 (Alan Liss, New York; 1992).
Flynn, T.G., Shyres, J. & Walton, D.J. Properties of the nicotinamide adenine dinucleotide phosphate dependent aldehyde reductase from pig kidney. J. biol. Chem. 250, 2933–2940 (1975).
Feldman, H.B. et al. Stereospecificity of the hydrogen transfer catalyzed by human placental aldose reductase. Biochim. biophys. Acta. 480, 14–20 (1977).
Bohren, K.M. et al Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: enzyme kinetics and crystal structure of the Y48H mutant enzyme. Biochemistry 3, 2021–2032 (1994).
Tarle, I., Borhani, D.W., Wilson, D.K., Quiocho, F.A. & Petrash, J.M. Probing the active site of human aldose reductase: site directed mutagenesis of Asp-43, Tyr-48, Lys-77 and His-110. J. biol. Chem. 268, 25687–25693 (1993).
Lui, S.-Q., Bhatnagar, A. & Srivastava, S.K. Bovine lens aldose reductase: pH dependance of steady-state kinetic parameters and nucleotide binding. J. biol. Chem. 268, 25494–25499 (1993).
Ring, M. & Huber, R.E. Multiple replacements establish the importance of tyrosine-503 in β-galactosidase (Escherichia coli). Arch. biochem. Biophys. 283, 342–350 (1990).
Kuliopulos, A., Mildvan, A.S., Shortle, D. & Talalay, P. Kinetic and ultraviolet spectroscopic studies of active-site mutants of Δ-3-ketosteroid isomerase. Biochemistry 28, 149–159 (1989).
Kuliopulos, A., Mullen, G.P., Xue, L. & Mildvan, A.S. Stereochemistry of the concerted enolization catalyzed by Δ-3-ketosteroid isomerase. Biochemistry 30, 3169–3178 (1991).
Davidson, W.S. & Flynn, T.G. Kinetics and mechanism of aldehyde reductase from pig kidney. Biochem. J. 177, 595–601 (1979).
Grimshaw, C.E. Aldose reductase: model for a new paradigm of enzymic perfection in detoxification catalysts. Biochemistry 31, 10139–10145 (1992).
Kubiseski, T.J. & Flynn, T.G. Studies on aldose reductase: probing the role of R268 in human aldose reductase by site directed mutagenesis. J. biol. Chem. 270, 16911–16917.
Wilson, D.K., Tarle, I., Petrash, J.M. & Quiocho, F.A. Refined 1.8 Å structure of human aldose reductase complexed with the potent inhibitor zopolrestat. Proc. natn. Acad. Sci. U.S.A. 90, 9847–9851 (1993).
Feather, M.S., Flynn, T.G., Munro, K.A., Kubiseski, T.J. & Walton, D.J. Catalysis of reduction of carbohydrate 2-oxoaldehydes (osones) by mammalian aldose reductase and aldehyde reductase. Biochim. biophys. Acta., in the press.
Cromlish, J.A. & Flynn, T.G. Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and with low Km aldehyde reductase of pig brain and pig kidney. J. biol. Chem. 258, 3583–3586 (1983).
McPherson, A. Crystallization of macromolecules: general principiles. Meth. Enzymol. 114, 112–120 (1985).
Messerschmidt, A. & Pflugrath, J.W. Crystal orientation and x-ray pattern prediction routines for area-detector diffractometer systems in macromolecular crystallography. J. appl. Crystallogr. 20, 306–315 (1987).
Hammersley, A.P., Svensson, S.O. & Thompson, A. Calibration and correction of spatial distortions in 2D detector systems. Nucl. Instr. Meth. A346, 312–322 (1994).
Otwinowski, Z. in Proc. CCP4 Study Weekend, 29–30 Jan 1991. Data collection and processing (Eds Sawyer, L., Issacs, N. & Burley, S.) 56–62 (SERC Daresbury Laboratory, U.K; 1993).
Collaborative computational project, number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D50, 760–763 (1994).
Brünger, A.T., Krukowski, A. & Erickson, J. Slow-cooling protocol for crystallographic refinement by simulated annealing. Acta. Crystallogr. A46, 585–593 (1990).
Jones, A.T. A graphics model building and refinement system for macromolecules. J. Appl. Cryst. 11, 268–272 (1978).
Luzzati, V. Traitement statistique des erreurs dans las détérmination des structures cristallines. Acta Crystallogr. 5, 802–810 (1952).
Carson, M. & Bugg, C.E. Algorithm for ribbon models of proteins. J. molec. Graphics 4, 121–122 (1986).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
El-Kabbani, O., Judge, K., Ginell, S. et al. Structure of porcine aldehyde reductase holoenzyme. Nat Struct Mol Biol 2, 687–692 (1995). https://doi.org/10.1038/nsb0895-687
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0895-687
This article is cited by
-
All in the family
Nature Structural & Molecular Biology (1995)