Abstract
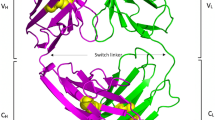
The pleckstrin homology (PH) ___domain is a conserved module present in many signal transducing and cytoskeletal proteins. Here we report the 2.8 Å crystal structure of the PH ___domain from dynamin. This ___domain consists of seven β-strands forming two roughly orthogonal antiparallel β-sheets terminating with an amphipathic α-helix. The structure also reveals a non-covalent dimeric association of the PH ___domain and a hydrophobic pocket surrounded by a charged rim. The dynamin PH ___domain structure is discussed in relation to its potential role in mediating interactions between proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Mayer, B.J., Ren, B.J., Clark, K.L. & Baltimore, D. A putative modular ___domain present in diverse signaling proteins. Cell 73, 629–630(1993).
Haslam, R.J., Koide, H.B. & Hemmings, B.A. Pleckstrin ___domain homology. Nature 363, 309–310 (1993).
Musacchio, A., Gibson, T., Rise, P., Thompson, J. & Saraste, M. The PH ___domain - a common piece in the structural patchwork of signaling proteins. Trends biochem. Sci., 343–348 (1993).
Shaw, G. Identification of novel pleckstrin homology (PH) domains provides a hypothesis for PH ___domain function. Biochem. biophys. Res. Commun. 195, 1145–1151 (1993).
Koch, W.J., Inglese, J., Stone, W.C. & Lefkowitz, R.J. The binding site for the β,γ-subunits of heterotrimetic G-proteins on the β-adrenergic-receptor kinase. J. biol. Chem. 268, 8256–8260 (1993).
Touhara, K., Inglese, J., Pitcher, J.A., Shaw, G. & Lefkowitz, R.J. Binding of G-protein β,γ-subunits to pleckstrin homology domains. J. biol. Chem. 269, 10217–10220 (1993).
Davis, L.H. & Bennett, V. Identification of 2 regions of β-spectrin that bind to distinct sites in brain membranes. J. biol. Chem. 269, 4409–4416 (1994).
Yoon, H.S., Hajduk, P.J., Petros, A.M., Olejniczak, E.T., Meadows, R.P. & Fesik, S.W. Solution structure of a pleckstrin-homology ___domain. Nature 369, 672–675 (1994).
Macias, M.J., Musacchio, A., Ponstingl, H., Nilges, M., Saraste, M. & Oschkinat, H. Structure of the pleckstrin homology ___domain from β- spectrin. Nature 369, 675–677 (1994).
van der Bliek, A.M., Redelmeier, T.E., Damke, H., Tisdale, E.J., Meyerowitz, E.M. & Schmid, S.L. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J. cell Biol. 122, 553–563 (1993).
Herskovits, J.S., Burgess, C.C., Obar, R.A. & Vallee, R.B., Effects of mutant rat dynamin on endocytosis. J. cell Biol. 122, 565–578 (1993).
Shpetner, H.S. & Valle, R.B. Dynamin is a GTPase stimulated to high levels of activity by microtubules. Nature 355, 733–735 (1992).
Ando, A. et al. A complex of Grb2 dynamin binds to tyrosine phosphorylated insulin-receptor substrate-after insulin-treatment. EMBO J. 13, 3033–3038 (1994).
Gout, I. et al. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell 75, 25–36 (1993).
Thomas, I.D., Sideras, P., Smith, C.I.E., Vorechovsky, I., Chapman, V. & Paul, W.E. Colocalization of X-linked agammaglobulinemia and X-linked immunodeficiency genes. Science 261, 355–358 (1993).
Rawlings, D.I. et al. Mutation of a unique region of Brutons tyrosine kinase in immunodeficient xid mice. Science 261, 358–361 (1993).
Blundell, T.L. & Johnson, L.N. in Protein crystallography (Academic, London; 1976).
Cowan, S.W., Newcomer, M.E. & Jones, T.A. Crystallographic refinement of human serum retinol binding-protein at 2Å resolution. Proteins. 8, 44–61 (1990).
Sali, A. & Blundell, T.L. The definition of topological equivalence in homologous and analogous structures: a procedure involving a comparison of local properties and relationships. J. molec. Biol. 212, 403–442 (1990).
Harlan, J.E., Hajduk, P.J., Yoon, H.S. & Fesik, S.W. Pleckstrin homology domains bind to phosphatidylinositol -4,5 - bisphosphate. Nature. 371, 168–170 (1994).
Pawson, T. & Schlessinger, J. SH2 and SH3 domains. Current Biol. 3, 434–441 (1993).
Simonds, W.F., Manji, H.K., Garritsen, A., Lupas, A.N. G-proteins and bark - a new twist for the coiled-coil. Trends biochem. Sci. 18, 315–317 (1993).
Ellis, M.V., Carne, A. & Katan, M. Structural requirements of phosphatidylinositol-specific phospholipase-cδ for enzyme activity. Eur J. Biochem. 213, 339–347 (1993).
Leslie, A.G.W. in Crystallographic Computing (Oxford Univ. Press; 1990).
CCP4 Collaborative Computing Project 4 (Daresbury Lab Warrington, UK; 1992).
Jones, T.A., Zou, J.-Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the ___location of errors in these models. Acta crystallogr. A47, 110–119 (1991).
Driessen, H., Haneef, M.I.J., Harris, G.W., Howlin, B., Khan, G. & Moss, D.S. Restrain: restrained structure-factor least-squares refinement program for macromolecular structures. J. appl. Crystallogr. 22, 510–516 (1989).
Brunger, A.T. X-PLOR Version 3.1 Manual (Yale Univ. Press, New Haven; 1992).
Evans, S.V. SETOR: hardware-lighted three-dimensional solid model representations of marcomolecules. J. molec. Graphics 11, 134–138 (1993).
Hutchinson, E.G. & Thornton, J.M. Hera - a program to draw schematic diagrams of protein secondary structures. Proteins 8, 203–212 (1990).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure. Pattern recognition of hydrogen bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Overington, J.R., Johnson, M.S., Sali, A. & BIundell, T.L. Tertiary structural constraints on protein evolutionary diversity: templates, key residues and structure prediction. Proc. Royal Soc. B241, 132–145 (1990).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Timm, D., Salim, K., Gout, I. et al. Crystal structure of the pleckstrin homology ___domain from dynamin. Nat Struct Mol Biol 1, 782–788 (1994). https://doi.org/10.1038/nsb1194-782
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb1194-782
This article is cited by
-
Crystal structure of nucleotide-free dynamin
Nature (2011)
-
The crystal structure of dynamin
Nature (2011)
-
Three-dimensional reconstruction of dynamin in the constricted state
Nature Cell Biology (2001)
-
Three ways to make a vesicle
Nature Reviews Molecular Cell Biology (2000)
-
Structural basis for IL-4 receptor phosphopeptide recognition by thelRS-1 PTB ___domain
Nature Structural & Molecular Biology (1996)