Abstract
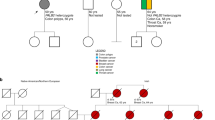
PALB2 loss-of-function variants play an important role in breast, pancreatic and possibly, ovarian and gastric cancer susceptibility. Their frequency can be influenced by founder effects, already described in some populations. Herein, we have assessed the possible founder effect of PALB2 c.2257C>T (p.Arg753*) truncating variant among Greek breast cancer patients, while investigating possible correlations with cancer diagnoses. Following a lead deriving from a background study of highly selected Greek breast cancer patients, a total of 2496 breast and 697 ovarian cancer patients were directly genotyped for the PALB2 c.2257C>T truncating variant. Consequently, haplotype analysis was conducted on identified carriers, using seven microsatellite markers. The prevalence of the PALB2 variant was 0.24% (6/2496) and 0.14% (1/697) among breast and ovarian cases, respectively. Family history seems to be an important factor for the variant identification, although not reaching statistical significance. Microsatellite analysis on 12 carriers revealed a common shared haplotype, spanning a chromosomal region of ~1.2 Mb; the variant was possibly introduced in the Greek population ~1600 years ago. The variant confers high breast cancer risk, as illustrated by comparison with publicly available control groups. Genetic testing for PALB2, especially for the Greek founder c.2257C>T truncating variant, should be seriously considered in Greek breast cancer cases, since such findings could assist appropriate clinical management for the patients and their families.
Similar content being viewed by others
Login or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–4.
Buisson R, Masson JY. PALB2 self-interaction controls homologous recombination. Nucleic Acids Res. 2012;40:10312–23.
Fernandes PH, Saam J, Peterson J, Hughes E, Kaldate R, Cummings S, et al. Comprehensive sequencing of PALB2 in patients with breast cancer suggests PALB2 mutations explain a subset of hereditary breast cancer. Cancer. 2014;120:963–7.
Tischkowitz M, Xia B, Sabbaghian N, Reis-Filho JS, Hamel N, Li G, et al. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci USA. 2007;104:6788–93.
Tischkowitz M, Xia B. PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res. 2010;70:7353–9.
Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7.
Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkas K, Roberts J, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506.
Lu HM, Li S, Black MH, Lee S, Hoiness R, Wu S, et al. Association of breast and ovarian cancers with predisposition genes identified by large-scale sequencing. JAMA Oncol. 2018. https://doi.org/10.1001/jamaoncol.2018.2956.
Couch FJ, Shimelis H, Hu C, Hart SN, Polley EC, Na J, et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017;3:1190–6.
Hauke J, Horvath J, Gross E, Gehrig A, Honisch E, Hackmann K, et al. Gene panel testing of 5589 BRCA1/2-negative index patients with breast cancer in a routine diagnostic setting: results of the german consortium for hereditary breast and ovarian cancer. Cancer Med. 2018;7:1349–58.
Castera L, Harter V, Muller E, Krieger S, Goardon N, Ricou A, et al. Landscape of pathogenic variations in a panel of 34 genes and cancer risk estimation from 5131 HBOC families. Genet Med. 2018;20:1677–86.
Ramus SJ, Song H, Dicks E, Tyrer JP, Rosenthal AN, Intermaggio MP, et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst. 2015. https://doi.org/10.1093/jnci/djv214.
Kotsopoulos J, Sopik V, Rosen B, Fan I, McLaughlin JR, Risch H, et al. Frequency of germline PALB2 mutations among women with epithelial ovarian cancer. Fam Cancer. 2017;16:29–34.
Norquist BM, Harrell MI, Brady MF, Walsh T, Lee MK, Gulsuner S, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2:482–90.
Zhen DB, Rabe KG, Gallinger S, Syngal S, Schwartz AG, Goggins MG, et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: a PACGENE study. Genet Med. 2015;17:569–77.
Borecka M, Zemankova P, Vocka M, Soucek P, Soukupova J, Kleiblova P, et al. Mutation analysis of the PALB2 gene in unselected pancreatic cancer patients in the Czech Republic. Cancer Genet. 2016;209:199–204.
Fewings E, Larionov A, Redman J, Goldgraben MA, Scarth J, Richardson S, et al. Germline pathogenic variants in PALB2 and other cancer-predisposing genes in families with hereditary diffuse gastric cancer without CDH1 mutation: a whole-exome sequencing study. Lancet Gastroenterol Hepatol. 2018;3:489–98.
Pritzlaff M, Summerour P, McFarland R, Li S, Reineke P, Dolinsky JS, et al. Male breast cancer in a multi-gene panel testing cohort: insights and unexpected results. Breast Cancer Res Treat. 2017;161:575–86.
Fostira F, Saloustros E, Apostolou P, Vagena A, Kalfakakou D, Mauri D, et al. Germline deleterious mutations in genes other than BRCA2 are infrequent in male breast cancer. Breast Cancer Res Treat. 2018;169:105–13.
Erkko H, Dowty JG, Nikkila J, Syrjakoski K, Mannermaa A, Pylkas K, et al. Penetrance analysis of the PALB2 c.1592delT founder mutation. Clin Cancer Res. 2008;14:4667–71.
Foulkes WD, Ghadirian P, Akbari MR, Hamel N, Giroux S, Sabbaghian N, et al. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007;9:R83.
Catucci I, Casadei S, Ding YC, Volorio S, Ficarazzi F, Falanga A, et al. Haplotype analyses of the c.1027C>T and c.2167_2168delAT recurrent truncating mutations in the breast cancer-predisposing gene PALB2. Breast Cancer Res Treat. 2016;160:121–9.
Southey MC, Teo ZL, Dowty JG, Odefrey FA, Park DJ, Tischkowitz M, et al. A PALB2 mutation associated with high risk of breast cancer. Breast Cancer Res. 2010;12:R109.
Fostira F, Walsh T, Casadei S, Lee MK, Vratimos A, Fountzilas G, et al., Combination of founder mutation screening and genomic capture using BROCA yield high rate of Loss-of-Function mutation in early onset and familial breast and ovarian cancer in Greece. ASHG meeting abstracts; 2013.
Reeve JP, Rannala B. DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics. 2002;18:894–5.
Papi L, Putignano AL, Congregati C, Piaceri I, Zanna I, Sera F, et al. A PALB2 germline mutation associated with hereditary breast cancer in Italy. Fam Cancer. 2010;9:181–5.
Hellebrand H, Sutter C, Honisch E, Gross E, Wappenschmidt B, Schem C, et al. Germline mutations in the PALB2 gene are population specific and occur with low frequencies in familial breast cancer. Hum Mutat. 2011;32:E2176–88.
Kim H, Cho DY, Choi DH, Oh M, Shin I, Park W, et al. Frequency of pathogenic germline mutation in CHEK2, PALB2, MRE11, and RAD50 in patients at high risk for hereditary breast cancer. Breast Cancer Res Treat. 2017;161:95–102.
Suszynska M, Klonowska K, Jasinska AJ, Kozlowski P. Large-scale meta-analysis of mutations identified in panels of breast/ovarian cancer-related genes - Providing evidence of cancer predisposition genes. Gynecol Oncol. 2019;153:452–62.
Hauke J, Horvath J, Gross E, Gehrig A, Honisch E, Hackmann K, et al. Gene panel testing of 5589 BRCA1/2-negative index patients with breast cancer in a routine diagnostic setting: results of the German Consortium for Hereditary Breast and Ovarian Cancer. Cancer Med. 2018;7:1349–58.
Buys SS, Sandbach JF, Gammon A, Patel G, Kidd J, Brown KL, et al. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer. 2017;123:1721–30.
Erkko H, Xia B, Nikkila J, Schleutker J, Syrjakoski K, Mannermaa A, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–9.
Girard E, Eon-Marchais S, Olaso R, Renault AL, Damiola F, Dondon MG, et al. Familial breast cancer and DNA repair genes: Insights into known and novel susceptibility genes from the GENESIS study, and implications for multigene panel testing. Int J Cancer. 2018. https://doi.org/10.1002/ijc.31921.
Slavin TP, Maxwell KN, Lilyquist J, Vijai J, Neuhausen SL, Hart SN, et al. The contribution of pathogenic variants in breast cancer susceptibility genes to familial breast cancer risk. NPJ Breast Cancer. 2017;3:22.
Shimelis H, LaDuca H, Hu C, Hart SN, Na J, Thomas A, et al. Triple-negative breast cancer risk genes identified by multigene hereditary cancer panel testing. J Natl Cancer Inst. 2018. https://doi.org/10.1093/jnci/djy106.
Nguyen-Dumont T, Hammet F, Mahmoodi M, Tsimiklis H, Teo ZL, Li R, et al. Mutation screening of PALB2 in clinically ascertained families from the breast cancer family registry. Breast Cancer Res Treat. 2015;149:547–54.
Couch FJ, Hart SN, Sharma P, Toland AE, Wang X, Miron P, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol. 2015;33:304–11.
Acknowledgements
We thank the patients and their families who participated in this study, as well as the clinicians for their valuable contribution on this project. We would also like to thank Dr. Laura Papi (Department of Experimental and Clinical Biomedical Sciences “Mario Serio”, Medical Genetics Unit, University of Florence, Florence, Italy) and her colleagues for providing DNA samples. Finally, we would like to thank Prof. Roberto Colombo (Faculty of Medicine “Agostino Gemelli”, Catholic University of the Sacred Heart, Rome, Italy) for his useful suggestions regarding haplotyping and microsatellite marker selection.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Vagena, A., Papamentzelopoulou, M., Kalfakakou, D. et al. PALB2 c.2257C>T truncating variant is a Greek founder and is associated with high breast cancer risk. J Hum Genet 64, 767–773 (2019). https://doi.org/10.1038/s10038-019-0612-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0612-6
This article is cited by
-
Two recurrent pathogenic/likely pathogenic variants in PALB2 account for half of PALB2 positive families in Slovenia
Human Genomics (2024)
-
Spectrum and characteristics of germline PALB2 pathogenic variants in 1556 early-onset breast cancer patients in China
Journal of Cancer Research and Clinical Oncology (2024)
-
Frequency of heterozygous germline pathogenic variants in genes for Fanconi anemia in patients with non-BRCA1/BRCA2 breast cancer: a meta-analysis
Breast Cancer Research and Treatment (2020)
-
Germline pathogenic variants in BRCA1, BRCA2, PALB2 and RAD51C in breast cancer women from Argentina
Breast Cancer Research and Treatment (2019)