Abstract
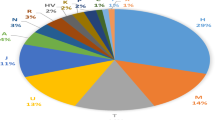
The study of mitochondrial DNA (mtDNA) variability at the level of whole mitogenomes has significant implications for the fields of human evolution and population genetics. In this paper, we present the results of a study of the complete mtDNA variability in Belarusians from the southeastern part of the Republic of Belarus. It was found that Southeast Belarusians are characterized by a high diversity of mitochondrial genomes. The analysis of genetic distances between European populations showed significant differences between the studied Belarusian sample from the bulk of East European populations, including Slavic ethnic groups. The results of the phylogeographic analysis indicated the presence of the West Asian component (12.6%) in the Belarusian mitochondrial gene pool, which can account for the observed genetic differences between Belarusians and other Eastern Slavs (Russians and Ukrainians). The East Asian component of the mitochondrial gene pool of the studied group of Belarusians is represented by haplogroup C5c1a (2.3%). The results of the phylogeographic analysis indicated that this mtDNA subclade is predominantly present in the gene pools of Slavic peoples, including Poles, Belarusians, Ukrainians, and Russians. The evolutionary age of haplogroup C5c1a is ~4000 years and, consequently, the appearance of C5c1-haplotypes in the eastern regions of Europe may be linked to the migrations of the Caspian steppe populations to the west during the Bronze Age.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Sedov VV. Slavs in the early Middle Ages. Moscow: Institute of Archaeology of RAS; 1995.
Barford PM. The early Slavs: culture and society in early medieval Eastern Europe. Ithaca, N.Y.: Cornell University Press; 2001.
Rębała K, Mikulich AI, Tsybovsky IS, Siváková D, Džupinková Z, Szczerkowska-Dobosz A, et al. Y-STR variation among Slavs: evidence for the Slavic homeland in the middle Dnieper basin. J Hum Genet. 2007;52:406–14. https://doi.org/10.1007/s10038-007-0125-6
Kushniarevich A, Utevska O, Chuhryaeva M, Agdzhoyan A, Dibirova K, Uktveryte I, et al. Genetic Heritage of the Balto-Slavic Speaking Populations: A Synthesis of Autosomal, Mitochondrial and Y-Chromosomal Data. PLoS One. 2015;10:e0135820. https://doi.org/10.1371/journal.pone.0135820
Malyarchuk B, Skonieczna K, Duleba A, Derenko M, Malyarchuk A, Grzybowski T. Mitogenomic diversity in Czechs and Slovaks. Forensic Sci Int Genet. 2022;59:102714. https://doi.org/10.1016/j.fsigen.2022.102714
Belyaeva O, Bermisheva M, Khrunin A, Slominsky P, Bebyakova N, Khusnutdinova E, et al. Mitochondrial DNA variations in Russian and Belorussian populations. Hum Biol. 2003;75:647–60
Kushniarevich A, Sivitskaya L, Danilenko N, Novogrodskii T, Tsybovsky I, Kiseleva A, et al. Uniparental genetic heritage of belarusians: encounter of rare middle eastern matrilineages with a central European mitochondrial DNA pool. PLoS One. 2013;8:e66499. https://doi.org/10.1371/journal.pone.0066499
Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, et al. Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet. 2001;69:1348–56. https://doi.org/10.1086/324511
Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. https://doi.org/10.1038/13779
Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2. https://doi.org/10.1093/bioinformatics/btp187
Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–7. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Malyarchuk B, Litvinov A, Derenko M, Skonieczna K, Grzybowski T, Grosheva A, et al. Mitogenomic diversity in Russians and Poles. Forensic Sci Int Genet. 2017;30:51–6. https://doi.org/10.1016/j.fsigen.2017.06.003
Malyarchuk B, Derenko M. Mitochondrial gene pool of Ukrainians in the context of variability of whole mitogenomes in Slavic peoples. Russ J Genet. 2023;59:88–96. https://doi.org/10.1134/S1022795423010088
Davidovic S, Malyarchuk B, Grzybowski T, Aleksic JM, Derenko M, Litvinov A, et al. Complete mitogenome data for the Serbian population: the contribution to high-quality forensic databases. Int J Leg Med. 2020;134:1581–90. https://doi.org/10.1007/s00414-020-02324-x
Malyarchuk B, Derenko M, Denisova G, Litvinov A, Rogalla U, Skonieczna K, et al. Whole mitochondrial genome diversity in two Hungarian populations. Mol Genet Genom. 2018;293:1255–63. https://doi.org/10.1007/s00438-018-1458
Malyarchuk B, Derenko M, Denisova G, Kravtsova O. Mitogenomic diversity in Tatars from the Volga-Ural region of Russia. Mol Biol Evol. 2010;27:2220–6. https://doi.org/10.1093/molbev/msq065
Stoljarova M, King JL, Takahashi M, Aaspõllu A, Budowle B. Whole mitochondrial genome genetic diversity in an Estonian population sample. Int J Leg Med. 2016;130:67–71. https://doi.org/10.1007/s00414-015-1249-4
Raule N, Sevini F, Li S, Barbieri A, Tallaro F, Lomartire L, et al. The co-occurrence of mtDNA mutations on different oxidative phosphorylation subunits, not detected by haplogroup analysis, affects human longevity and is population specific. Aging Cell. 2014;13:401–7. https://doi.org/10.1111/acel.12186
Fraumene C, Belle EM, Castrì L, Sanna S, Mancosu G, Cosso M, et al. High-resolution analysis and phylogenetic network construction using complete mtDNA sequences in Sardinian genetic isolates. Mol Biol Evol. 2006;23:2101–11. https://doi.org/10.1093/molbev/msl084
Fan L, Yao YG. An update to MitoTool: using a new scoring system for faster mtDNA haplogroup determination. Mitochondrion. 2013;13:360–3. https://doi.org/10.1016/j.mito.2013.04.011
van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:386–94. https://doi.org/10.1002/humu.20921
Dür A, Huber N, Parson W. Fine-tuning phylogenetic alignment and haplogrouping of mtDNA sequences. Int J Mol Sci. 2021;22:5747. https://doi.org/10.3390/ijms22115747
Mallick S, Micco A, Mah M, Ringbauer H, Lazaridis I, Olalde I, et al. The Allen Ancient DNA Resource (AADR) is a curated compendium of ancient human genomes. Sci Data. 2024;11;182. https://doi.org/10.1038/s41597-024-03031-7
Soares P, Ermini L, Thomson N, Mormina M, Rito T, Röhl A, et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–59. https://doi.org/10.1016/j.ajhg.2009.05.001
Macaulay V, Soares P, Richards MB. Rectifying long-standing misconceptions about the ρ statistic for molecular dating. PLoS One. 2019;14:e0212311. https://doi.org/10.1371/journal.pone.0212311
Derenko M, Malyarchuk B, Bahmanimehr A, Denisova G, Perkova M, Farjadian S, et al. Complete mitochondrial DNA diversity in Iranians. PLoS One. 2013;8:e80673. https://doi.org/10.1371/journal.pone.0080673
Matisoo-Smith EA, Gosling AL, Boocock J, Kardailsky O, Kurumilian Y, Roudesli-Chebbi S, et al. PLoS One. 2016;11:e0155046. https://doi.org/10.1371/journal.pone.0155046
Peng MS, Xu W, Song JJ, Chen X, Sulaiman X, Cai L, et al. Mitochondrial genomes uncover the maternal history of the Pamir populations. Eur J Hum Genet. 2018;26:124–36. https://doi.org/10.1038/s41431-017-0028-8
Derenko M, Denisova G, Malyarchuk B, Hovhannisyan A, Khachatryan Z, Hrechdakian P, et al. Insights into matrilineal genetic structure, differentiation and ancestry of Armenians based on complete mitogenome data. Mol Gen Genom. 2019;294:1547–59. https://doi.org/10.1007/s00438-019-01596-2
Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, et al. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet. 2000;67:1251–76
Narasimhan VM, Patterson N, Moorjani P, Rohland N, Bernardos R, Mallick S, et al. The formation of human populations in South and Central Asia. Science. 2019;365:eaat7487 https://doi.org/10.1126/science.aat7487
Krzewińska M, Kılınç GM, Juras A, Koptekin D, Chyleński M, Nikitin AG, et al. Ancient genomes suggest the eastern Pontic-Caspian steppe as the source of western Iron Age nomads. Sci Adv. 2018;4:eaat4457. https://doi.org/10.1126/sciadv.aat4457
Maróti Z, Neparáczki E, Schütz O, Maár K, Varga GIB, Kovács B, et al. The genetic origin of Huns, Avars, and conquering Hungarians. Curr Biol. 2022;32:2858–70.e7. https://doi.org/10.1016/j.cub.2022.04.093
Szeifert B, Gerber D, Csáky V, Langó P, Stashenkov DA, Khokhlov AA, et al. Tracing genetic connections of ancient Hungarians to the 6th-14th century populations of the Volga-Ural region. Hum Mol Genet. 2022;31:3266–80. https://doi.org/10.1093/hmg/ddac106
Vyas DN, Koncz I, Modi A, Mende BG, Tian Y, Francalacci P, et al. Fine-scale sampling uncovers the complexity of migrations in 5th-6th century Pannonia. Curr Biol. 2023;33:3951–61.e11. https://doi.org/10.1016/j.cub.2023.07.063
Gnecchi-Ruscone GA, Rácz Z, Samu L, Szeniczey T, Faragó N, Knipper C, et al. Network of large pedigrees reveals social practices of Avar communities. Nature. 2024;629:376–83. https://doi.org/10.1038/s41586-024-07312-4
Pankratov V, Litvinov S, Kassian A, Shulhin D, Tchebotarev L, Yunusbayev B, et al. East Eurasian ancestry in the middle of Europe: genetic footprints of Steppe nomads in the genomes of Belarusian Lipka Tatars. Sci Rep. 2016;6:30197. https://doi.org/10.1038/srep30197.
Gnecchi-Ruscone GA, Szécsényi-Nagy A, Koncz I, Csiky G, Rácz Z, Rohrlach AB, et al. Ancient genomes reveal origin and rapid trans-Eurasian migration of 7th century Avar elites. Cell. 2022;185:1402–13.e21. https://doi.org/10.1016/j.cell.2022.03.007
Płoszaj T, Jędrychowska-Dańska K, Zamerska A, Drozd-Lipińska A, Poliński D, Janowski A, et al. Ancient DNA analysis might suggest external origin of individuals from chamber graves placed in medieval cemetery in Pień, Central Poland. Anthropol Anz. 2017;74:319–37. https://doi.org/10.1127/anthranz/2017/0727
Acknowledgements
We thank all the donors and Dr. I.S. Tsybovsky for providing samples for this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Malyarchuk, B., Denisova, G. & Litvinov, A. Heterogeneity of the Southeast Belarusian mitochondrial gene pool. J Hum Genet 70, 313–320 (2025). https://doi.org/10.1038/s10038-025-01337-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-025-01337-x