Abstract
Background
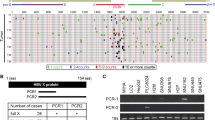
Hepatoblastoma (HB) is a highly aggressive paediatric malignancy that exhibits a high presence of cancer stem cells (CSCs), which related to tumour recurrence and chemotherapy resistance. Brain expressed X-linked protein 1 (BEX1) plays a pivotal role in ciliogenesis, axon regeneration and differentiation of neural stem cells. However, the role of BEX1 in metabolic and stemness programs in HB remains unclear.
Methods
BEX1 expression in human and mouse HB was analyzed using gene expression profile data from NCBI GEO and immunohistochemical validation. Seahorse extracellular flux analyzer, ultra-high-performance liquid-chromatography mass spectrometry (LC-MS), flow cytometry, qRT-PCR, Western Blot, sphere formation assay, and diluted xenograft tumour formation assay were used to analyze metabolic and stemness features.
Results
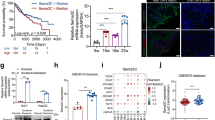
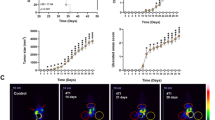
Our results indicated that overexpression of BEX1 significantly enhanced the Warburg effect in HB cells. Furthermore, glycolysis inhibition largely attenuated the effects of BEX1 on HB cell growth and self-renewal, suggesting that BEX1 promotes stemness maintenance of HB cells by regulating the Warburg effect. Mechanistically, BEX1 enhances Warburg effect through the downregulation of peroxisome proliferator-activated receptor-gamma (PPARγ). Furthermore, pyruvate dehydrogenase kinase isozyme 1 (PDK1) is required for PPARγ-induced inhibition of Warburg effect in HB. In addition, BEX1 supports the stemness of HB by enhancing Warburg effect in a PPARγ/PDK1 dependent manner.
Conclusions
HB patients with high BEX1 and PDK1 expression had a poor prognosis. BEX1 promotes the stemness maintenance of HB cells via modulating the Warburg effect, which depends on PPARγ/PDK1 axis. Pioglitazone could be used to target BEX1-mediated stemness properties in HB by upregulating PPARγ.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
269,00 € per year
only 11,21 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All of the relevant data are included in supplemental information. RNA-seq data have been submitted to the SRA database (SRA: PRJNA721822). All data are available upon request to the corresponding authors.
References
Sumazin P, Peters TL, Sarabia SF, Kim HR, Urbicain M, Hollingsworth EF, et al. Hepatoblastomas with carcinoma features represent a biological spectrum of aggressive neoplasms in children and young adults. J Hepatol. 2022;77:1026–37.
Trobaugh-Lotrario A, Katzenstein HM, Ranganathan S, Lopez-Terrada D, Krailo MD, Piao J, et al. Small cell undifferentiated histology does not adversely affect outcome in hepatoblastoma: a report from the children’s oncology group (COG) AHEP0731 study committee. J Clin Oncol. 2022;40:459–67.
Loesch R, Caruso S, Paradis V, Godard C, Gougelet A, Renault G, et al. Deleting the beta-catenin degradation ___domain in mouse hepatocytes drives hepatocellular carcinoma or hepatoblastoma-like tumour growth. J Hepatol. 2022;77:424–35.
Semeraro M, Branchereau S, Maibach R, Zsiros J, Casanova M, Brock P, et al. Relapses in hepatoblastoma patients: clinical characteristics and outcome–experience of the International Childhood Liver Tumour Strategy Group (SIOPEL). Eur J Cancer. 2013;49:915–22.
Perilongo G, Shafford E, Plaschkes J. SIOPEL trials using preoperative chemotherapy in hepatoblastoma. Lancet Oncol. 2000;1:94–100.
Cairo S, Armengol C, De Reynies A, Wei Y, Thomas E, Renard CA, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–84.
Aronson DC, Czauderna P, Maibach R, Perilongo G, Morland B. The treatment of hepatoblastoma: its evolution and the current status as per the SIOPEL trials. J Indian Assoc Pediatr Surg. 2014;19:201–7.
Ortega JA, Douglass EC, Feusner JH, Reynolds M, Quinn JJ, Finegold MJ, et al. Randomized comparison of cisplatin/vincristine/fluorouracil and cisplatin/continuous infusion doxorubicin for treatment of pediatric hepatoblastoma: a report from the children’s cancer group and the pediatric oncology group. J Clin Oncol. 2000;18:2665–75.
Rougemont AL, McLin VA, Toso C, Wildhaber BE. Adult hepatoblastoma: learning from children. J Hepatol. 2012;56:1392–403.
Yang T, Liang N, Li J, Hu P, Huang Q, Zhao ZF, et al. MDSCs might be “Achilles heel” for eradicating CSCs. Cytokine Growth Factor Rev. 2022;65:39–50.
Carrillo-Reixach J, Torrens L, Simon-Coma M, Royo L, Domingo-Sabat M, Abril-Fornaguera J, et al. Epigenetic footprint enables molecular risk stratification of hepatoblastoma with clinical implications. J Hepatol. 2020;73:328–41.
Jagadisan B, Dhawan A. Emergencies in paediatric hepatology. J Hepatol. 2022;76:1199–214.
Liang N, Yang T, Huang Q, Yu P, Liu C, Chen L, et al. Mechanism of cancer stemness maintenance in human liver cancer. Cell Death Dis. 2022;13:394.
Marayati R, Stafman LL, Williams AP, Bownes LV, Quinn CH, Markert HR, et al. CRISPR/Cas9-mediated knockout of PIM3 suppresses tumourigenesis and cancer cell stemness in human hepatoblastoma cells. Cancer Gene Ther. 2022;29:558–72.
Mavila N, Thundimadathil J. The Emerging Roles of Cancer Stem Cells and Wnt/Beta-Catenin Signaling in Hepatoblastoma. Cancers (Basel). 2019;11:1406.
Monga SP. Beta-catenin signaling and roles in liver homeostasis, injury, and tumourigenesis. Gastroenterology. 2015;148:1294–310.
Russell JO, Monga SP. Wnt/beta-catenin signaling in liver development, homeostasis, and pathobiology. Annu Rev Pathol. 2018;13:351–78.
Wang H, Zhang S, Zhang Y, Jia J, Wang J, Liu X, et al. TAZ is indispensable for c-MYC-induced hepatocarcinogenesis. J Hepatol. 2022;76:123–34.
Wang G, Wang Q, Liang N, Xue H, Yang T, Chen X, et al. Oncogenic driver genes and tumour microenvironment determine the type of liver cancer. Cell Death Dis. 2020;11:313.
Sagawa H, Naiki-Ito A, Kato H, Naiki T, Yamashita Y, Suzuki S, et al. Connexin 32 and luteolin play protective roles in non-alcoholic steatohepatitis development and its related hepatocarcinogenesis in rats. Carcinogenesis. 2015;36:1539–49.
Gu Y, Wei W, Cheng Y, Wan B, Ding X, Wang H, et al. A pivotal role of BEX1 in liver progenitor cell expansion in mice. Stem Cell Res Ther. 2018;9:164.
Khazaei MR, Halfter H, Karimzadeh F, Koo JH, Margolis FL, Young P. Bex1 is involved in the regeneration of axons after injury. J Neurochem. 2010;115:910–20.
Koo JH, Smiley MA, Lovering RM, Margolis FL. Bex1 knock out mice show altered skeletal muscle regeneration. Biochem Biophys Res Commun. 2007;363:405–10.
de Ronde JJ, Lips EH, Mulder L, Vincent AD, Wesseling J, Nieuwland M, et al. SERPINA6, BEX1, AGTR1, SLC26A3, and LAPTM4B are markers of resistance to neoadjuvant chemotherapy in HER2-negative breast cancer. Breast Cancer Res Treat. 2013;137:213–23.
Foltz G, Ryu GY, Yoon JG, Nelson T, Fahey J, Frakes A, et al. Genome-wide analysis of epigenetic silencing identifies BEX1 and BEX2 as candidate tumour suppressor genes in malignant glioma. Cancer Res. 2006;66:6665–74.
Wang Q, Liang N, Yang T, Li Y, Li J, Huang Q, et al. DNMT1-mediated methylation of BEX1 regulates stemness and tumourigenicity in liver cancer. J Hepatol. 2021;75:1142–53.
Ding K, Su Y, Pang L, Lu Q, Wang Z, Zhang S, et al. Inhibition of apoptosis by downregulation of hBex1, a novel mechanism, contributes to the chemoresistance of Bcr/Abl+ leukemic cells. Carcinogenesis. 2009;30:35–42.
Doi T, Ogawa H, Tanaka Y, Hayashi Y, Maniwa Y. Bex1 significantly contributes to the proliferation and invasiveness of malignant tumour cells. Oncol Lett. 2020;20:362.
Hiyama E, Ueda Y, Kurihara S, Kawashima K, Ikeda K, Morihara N, et al. Gene expression profiling in hepatoblastoma cases of the Japanese study group for pediatric liver tumours-2 (JPLT-2) trial, Eur. J. Mol. Cancer. 2019;1:2–8.
Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10.
Wang G, Wang Q, Huang Q, Chen Y, Sun X, He L, et al. Upregulation of mtSSB by interleukin-6 promotes cell growth through mitochondrial biogenesis-mediated telomerase activation in colorectal cancer. Int J Cancer. 2019;144:2516–28.
Wang D, Tian J, Yan Z, Yuan Q, Wu D, Liu X, et al. Mitochondrial fragmentation is crucial for c-Myc-driven hepatoblastoma-like liver tumours. Mol Ther. 2022;30:1645–60.
Liu P, Ge M, Hu J, Li X, Che L, Sun K, et al. A functional mammalian target of rapamycin complex 1 signaling is indispensable for c-Myc-driven hepatocarcinogenesis. Hepatology. 2017;66:167–81.
Yun WJ, Shin E, Lee K, Jung HY, Kim SH, Park YN, et al. Clinicopathologic implication of hepatic progenitor cell marker expression in hepatoblastoma. Pathol Res Pract. 2013;209:568–73.
Wu JF, Ho MC, Ni YH, Hsu HY, Lee PH, Chang MH, et al. Dysregulation of liver developmental microRNA contribute to hepatic carcinogenesis. J Formos Med Assoc. 2020;119:1041–51.
Ward SC, Thung SN, Lim KH, Tran TT, Hong TK, Hoang PL, et al. Hepatic progenitor cells in liver cancers from Asian children. Liver Int. 2010;30:102–11.
Xu X, Liu RF, Zhang X, Huang LY, Chen F, Fei QL, et al. DLK1 as a potential target against cancer stem/progenitor cells of hepatocellular carcinoma. Mol Cancer Ther. 2012;11:629–38.
Falix FA, Aronson DC, Lamers WH, Hiralall JK, Seppen J. DLK1, a serum marker for hepatoblastoma in young infants. Pediatr Blood Cancer. 2012;59:743–5.
Ilmer M, Garnier A, Vykoukal J, Alt E, von Schweinitz D, Kappler R, et al. Targeting the Neurokinin-1 receptor compromises canonical Wnt signaling in hepatoblastoma. Mol Cancer Ther. 2015;14:2712–21.
Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y, et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469–83.
Zeng SS, Yamashita T, Kondo M, Nio K, Hayashi T, Hara Y, et al. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. J Hepatol. 2014;60:127–34.
Lee TK, Castilho A, Cheung VC, Tang KH, Ma S, Ng IO. CD24(+) liver tumour-initiating cells drive self-renewal and tumour initiation through STAT3-mediated NANOG regulation. Cell Stem Cell. 2011;9:50–63.
Wang R, Li Y, Tsung A, Huang H, Du Q, Yang M, et al. iNOS promotes CD24(+)CD133(+) liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc Natl Acad Sci USA. 2018;115:E10127–36.
Cao W, Li M, Liu J, Zhang S, Noordam L, Verstegen MMA, et al. LGR5 marks targetable tumour-initiating cells in mouse liver cancer. Nat Commun. 2020;11:1961.
Wei Z, Jia J, Heng G, Xu H, Shan J, Wang G, et al. Sirtuin-1/mitochondrial ribosomal protein S5 Axis enhances the metabolic flexibility of liver cancer stem cells. Hepatology. 2019;70:1197–213.
Wei RR, Zhang MY, Rao HL, Pu HY, Zhang HZ, Wang HY. Identification of ADH4 as a novel and potential prognostic marker in hepatocellular carcinoma. Med Oncol. 2012;29:2737–43.
Liu X, Li T, Kong D, You H, Kong F, Tang R. Prognostic implications of alcohol dehydrogenases in hepatocellular carcinoma. BMC Cancer. 2020;20:1204.
Liu Q, Li J, Zhang W, Xiao C, Zhang S, Nian C, et al. Glycogen accumulation and phase separation drives liver tumour initiation. Cell. 2021;184:5559–76.e5519.
Hong SM, Lee YK, Park I, Kwon SM, Min S, Yoon G. Lactic acidosis caused by repressed lactate dehydrogenase subunit B expression down-regulates mitochondrial oxidative phosphorylation via the pyruvate dehydrogenase (PDH)-PDH kinase axis. J Biol Chem. 2019;294:7810–20.
Antonowicz S, Bodai Z, Wiggins T, Markar SR, Boshier PR, Goh YM, et al. Endogenous aldehyde accumulation generates genotoxicity and exhaled biomarkers in esophageal adenocarcinoma. Nat Commun. 2021;12:1454.
Gao T, Zhang X, Zhao J, Zhou F, Wang Y, Zhao Z, et al. SIK2 promotes reprogramming of glucose metabolism through PI3K/AKT/HIF-1alpha pathway and Drp1-mediated mitochondrial fission in ovarian cancer. Cancer Lett. 2020;469:89–101.
Yu J, Shen B, Chu ES, Teoh N, Cheung KF, Wu CW, et al. Inhibitory role of peroxisome proliferator-activated receptor gamma in hepatocarcinogenesis in mice and in vitro. Hepatology. 2010;51:2008–19.
Picard F, Auwerx J. PPAR(gamma) and glucose homeostasis. Annu Rev Nutr. 2002;22:167–97.
Shashni B, Sakharkar KR, Nagasaki Y, Sakharkar MK. Glycolytic enzymes PGK1 and PKM2 as novel transcriptional targets of PPARgamma in breast cancer pathophysiology. J Drug Target. 2013;21:161–74.
Zhang W, Shao W, Dong Z, Zhang S, Liu C, Chen S. Cloxiquine, a traditional antituberculosis agent, suppresses the growth and metastasis of melanoma cells through activation of PPARgamma. Cell Death Dis. 2019;10:404.
Benit P, Pelhaitre A, Saunier E, Bortoli S, Coulibaly A, Rak M, et al. Paradoxical inhibition of glycolysis by pioglitazone opposes the mitochondriopathy caused by AIF deficiency. EBioMedicine. 2017;17:75–87.
Morris NL, Michael DN, Crotty KM, Chang SS, Yeligar SM. Alcohol-induced glycolytic shift in alveolar macrophages is mediated by hypoxia-inducible Factor-1 alpha. Front Immunol. 2022;13:865492.
Zhu Y, Ji JJ, Wang XD, Sun XJ, Li M, Wei Q, et al. Periostin promotes arterial calcification through PPARgamma-related glucose metabolism reprogramming. Am J Physiol Heart Circ Physiol. 2021;320:H2222–39.
Yang Y, Zhao LH, Huang B, Wang RY, Yuan SX, Tao QF, et al. Pioglitazone, a PPARgamma agonist, inhibits growth and invasion of human hepatocellular carcinoma via blockade of the rage signaling. Mol Carcinog. 2015;54:1584–95.
Uehara T, Ainslie GR, Kutanzi K, Pogribny IP, Muskhelishvili L, Izawa T, et al. Molecular mechanisms of fibrosis-associated promotion of liver carcinogenesis. Toxicol Sci. 2013;132:53–63.
Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33.
Chen JQ, Russo J. Dysregulation of glucose transport, glycolysis, TCA cycle and glutaminolysis by oncogenes and tumour suppressors in cancer cells. Biochim Biophys Acta. 2012;1826:370–84.
Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95.
Tamai K, Nakamura-Shima M, Shibuya-Takahashi R, Kanno SI, Yasui A, Mochizuki M, et al. BEX2 suppresses mitochondrial activity and is required for dormant cancer stem cell maintenance in intrahepatic cholangiocarcinoma. Sci Rep. 2020;10:21592.
Sancho P, Barneda D, Heeschen C. Hallmarks of cancer stem cell metabolism. Br J Cancer. 2016;114:1305–12.
Chen CL, Uthaya Kumar DB, Punj V, Xu J, Sher L, Tahara SM, et al. NANOG metabolically reprograms tumour-initiating stem-like cells through tumourigenic changes in oxidative phosphorylation and fatty acid metabolism. Cell Metab. 2016;23:206–19.
Shen YA, Wang CY, Hsieh YT, Chen YJ, Wei YH. Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle. 2015;14:86–98.
Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–31.
Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–41.
Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014;4:a014241.
Folmes CD, Martinez-Fernandez A, Faustino RS, Yamada S, Perez-Terzic C, Nelson TJ, et al. Nuclear reprogramming with c-Myc potentiates glycolytic capacity of derived induced pluripotent stem cells. J Cardiovasc Transl Res. 2013;6:10–21.
Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20:649–88.
Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, et al. MYC/PGC-1alpha balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015;22:590–605.
Janiszewska M, Suva ML, Riggi N, Houtkooper RH, Auwerx J, Clement-Schatlo V, et al. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012;26:1926–44.
Ye XQ, Li Q, Wang GH, Sun FF, Huang GJ, Bian XW, et al. Mitochondrial and energy metabolism-related properties as novel indicators of lung cancer stem cells. Int J Cancer. 2011;129:820–31.
Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, et al. PPARgamma signaling and metabolism: the good, the bad and the future. Nat Med. 2013;19:557–66.
Choi JH, Banks AS, Kamenecka TM, Busby SA, Chalmers MJ, Kumar N, et al. Antidiabetic actions of a non-agonist PPARgamma ligand blocking Cdk5-mediated phosphorylation. Nature. 2011;477:477–81.
Choi JH, Banks AS, Estall JL, Kajimura S, Bostrom P, Laznik D, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466:451–6.
Quan Q, Qian Y, Li X, Li M. Pioglitazone reduces beta amyloid levels via inhibition of PPARgamma phosphorylation in a neuronal model of Alzheimer’s disease. Front Aging Neurosci. 2019;11:178.
Poulsen L, Siersbaek M, Mandrup S. PPARs: fatty acid sensors controlling metabolism. Semin Cell Dev Biol. 2012;23:631–9.
van Beekum O, Fleskens V, Kalkhoven E. Posttranslational modifications of PPAR-gamma: fine-tuning the metabolic master regulator. Obesity (Silver Spring). 2009;17:213–9.
Hsu HT, Sung MT, Lee CC, Kuo YJ, Chi CW, Lee HC, et al. Peroxisome proliferator-activated receptor gamma expression is inversely associated with macroscopic vascular invasion in human hepatocellular carcinoma. Int J Mol Sci. 2016;17:1226.
Cao LQ, Shao ZL, Liang HH, Zhang DW, Yang XW, Jiang XF, et al. Activation of peroxisome proliferator-activated receptor-gamma (PPARgamma) inhibits hepatoma cell growth via downregulation of SEPT2 expression. Cancer Lett. 2015;359:127–35.
Roche TE, Baker JC, Yan X, Hiromasa Y, Gong X, Peng T, et al. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog Nucleic Acid Res Mol Biol. 2001;70:33–75.
Zuo Q, He J, Zhang S, Wang H, Jin G, Jin H, et al. PPARgamma coactivator-1alpha suppresses metastasis of hepatocellular carcinoma by inhibiting warburg effect by PPARgamma-dependent WNT/beta-Catenin/Pyruvate dehydrogenase kinase isozyme 1 Axis. Hepatology. 2021;73:644–60.
Bamodu OA, Chang HL, Ong JR, Lee WH, Yeh CT, Tsai JT. Elevated PDK1 expression drives PI3K/AKT/MTOR signaling promotes radiation-resistant and dedifferentiated phenotype of hepatocellular carcinoma. Cells. 2020;9:746.
Dupuy F, Tabaries S, Andrzejewski S, Dong Z, Blagih J, Annis MG, et al. PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab. 2015;22:577–89.
Baumunk D, Reichelt U, Hildebrandt J, Krause H, Ebbing J, Cash H, et al. Expression parameters of the metabolic pathway genes pyruvate dehydrogenase kinase-1 (PDK-1) and DJ-1/PARK7 in renal cell carcinoma (RCC). World J Urol. 2013;31:1191–6.
Koukourakis MI, Giatromanolaki A, Harris AL, Sivridis E. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumour-associated stroma. Cancer Res. 2006;66:632–7.
Wang XQ, Lo CM, Chen L, Ngan ES, Xu A, Poon RY. CDK1-PDK1-PI3K/Akt signaling pathway regulates embryonic and induced pluripotency. Cell Death Differ. 2017;24:38–48.
Ling LS, Voskas D, Woodgett JR. Activation of PDK-1 maintains mouse embryonic stem cell self-renewal in a PKB-dependent manner. Oncogene. 2013;32:5397–408.
Wang Z, Xu X, Liu N, Cheng Y, Jin W, Zhang P, et al. SOX9-PDK1 axis is essential for glioma stem cell self-renewal and temozolomide resistance. Oncotarget. 2018;9:192–204.
Peng F, Wang JH, Fan WJ, Meng YT, Li MM, Li TT, et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene. 2018;37:1062–74.
Acknowledgements
We thank Professor Xin Chen and Professor Perry Hackett for providing the plasmids. The schematic diagram in this article was drawn by Figdraw. We thank Ms. Jie Chen for her help in designing the schematic diagram.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 82103664), Joint Construction Project of Henan Medical Science and Technology (LHGJ20200281) and Guangzhou Basic and Applied Basic Research Project (No. 202102021274).
Author information
Authors and Affiliations
Contributions
Conception and design: GW, XH, TY, QW. Development of methodology: JL, Y, SL. Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): QW, QH, LS, LT. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): QW, CL, NL. Writing, review, and/or revision of the manuscript: GW, XH, TY. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): CZ, YT. Study supervision: GW, TY, QW, XH.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All animal experiments were approved by the Institutional Animal Care and Use Committee of Zhengzhou University in accordance with NIH guidelines.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Q., Liang, N., Liu, C. et al. BEX1 supports the stemness of hepatoblastoma by facilitating Warburg effect in a PPARγ/PDK1 dependent manner. Br J Cancer 129, 1477–1489 (2023). https://doi.org/10.1038/s41416-023-02418-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02418-4