Abstract
Background
Screening for BCR-ABL1 kinase ___domain (KD) mutations is routinely implemented in cases of treatment failure for chronic myeloid leukaemia and Philadelphia-positive acute lymphoblastic leukaemia. However, timely deciphering their clonal relationship via mutation profiling that requires identification of mutation types, quantification of mutant abundance, and differentiation between compound and polyclonal mutations (CMs and PMs), remains difficult during therapy.
Methods
Herein, we established a protocol that identified mutation types and further distinguished clonal relationships by combining mini-sequencing of MeltArray with allele segregation of droplet digital PCR (ddPCR).
Results
The analysis showed that 78 samples (18.93%) were mutant, of which 50 (64.1%) harboured single mutations, and 28 (35.9%) contained multiple mutations, including double-, triple-, quadruple- and hepta-mutants. These results agreed with NGS, except one sample with F317L and L324Q mutations, where L324Q was beyond MeltArray’s scope. Among cases containing multiple mutations, 85.71% were PMs, 10.71% were CMs, and 3.57% were mixed CMs and PMs. Retrospective analysis revealed that clonal relationships in BCR-ABL1 KD mutations were highly dynamic during therapy.
Conclusions
The MeltArray-ddPCR protocol enables dynamic profiling of BCR-ABL1 KD mutations to determine clonal status, improving prediction of drug susceptibility and leukaemia outcomes.

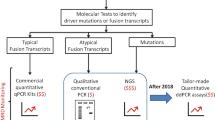
In this study, we performed a retrospective analysis of 539 samples from 365 leukaemia patients. We developed a comprehensive BCR-ABL1 kinase ___domain mutation screening protocol that includes pre-amplification, mutation screening, and differentiation between compound mutations (CMs) and polyclonal mutations (PMs). By integrating MeltArray and ddPCR technologies, this protocol enables dynamic monitoring of changes in mutation levels and clonal evolution. It also offers companion diagnostics based on mutational profiles to guide precision therapy for leukaemia patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
269,00 € per year
only 11,21 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
References
O’Hare T, Zabriskie MS, Eiring AM, Deininger MW. Pushing the limits of targeted therapy in chronic myeloid leukaemia. Nat Rev Cancer. 2012;12:513–26.
Deininger MW, Hodgson JG, Shah NP, Cortes JE, Kim DW, Nicolini FE, et al. Compound mutations in BCR-ABL1 are not major drivers of primary or secondary resistance to ponatinib in CP-CML patients. Blood. 2016;127:703–12.
Corbin AS, La Rosee P, Stoffregen EP, Druker BJ, Deininger MW. Several Bcr-Abl kinase ___domain mutants associated with imatinib mesylate resistance remain sensitive to imatinib. Blood. 2003;101:4611–4.
Eide CA, O’Hare T. Chronic myeloid leukemia: advances in understanding disease biology and mechanisms of resistance to tyrosine kinase inhibitors. Curr Hematol Malig Rep. 2015;10:158–66.
Kantarjian HM, Giles F, Gattermann N, Bhalla K, Alimena G, Palandri F, et al. Nilotinib (formerly AMN107), a highly selective BCR-ABL tyrosine kinase inhibitor, is effective in patients with Philadelphia chromosome-positive chronic myelogenous leukemia in chronic phase following imatinib resistance and intolerance. Blood. 2007;110:3540–6.
Zwaan CM, Rizzari C, Mechinaud F, Lancaster DL, Lehrnbecher T, van der Velden VH, et al. Dasatinib in children and adolescents with relapsed or refractory leukemia: results of the CA180-018 phase I dose-escalation study of the Innovative Therapies for Children with Cancer Consortium. J Clin Oncol. 2013;31:2460–8.
Gambacorti-Passerini C, Brummendorf TH, Kim DW, Turkina AG, Masszi T, Assouline S, et al. Bosutinib efficacy and safety in chronic phase chronic myeloid leukemia after imatinib resistance or intolerance: Minimum 24-month follow-up. Am J Hematol. 2014;89:732–42.
Khorashad JS, Kelley TW, Szankasi P, Mason CC, Soverini S, Adrian LT, et al. BCR-ABL1 compound mutations in tyrosine kinase inhibitor-resistant CML: frequency and clonal relationships. Blood. 2013;121:489–98.
Eide CA, Zabriskie MS, Adrian LT, Lange T, Deininger MW, Druker BJ, et al. Resistance profiling of BCR-ABL compound mutations linked to tyrosine kinase inhibitor therapy failure in chronic myeloid leukemia. Blood. 2011;118:1416–1416.
Zabriskie MS, Eide CA, Tantravahi SK, Vellore NA, Estrada J, Nicolini FE, et al. BCR-ABL1 compound mutations combining key kinase ___domain positions confer clinical resistance to ponatinib in Ph chromosome-positive leukemia. Cancer Cell. 2014;26:428–42.
Iqbal Z, Absar M, Mahmood A, Aleem A, Iqbal M, Jameel A, et al. Discovery and protein modeling studies of novel compound mutations causing resistance to multiple tyrosine kinase inhibitors in chronic myeloid leukemia. Asian Pac J Cancer Prev. 2020;21:3517–26.
Parker WT, Yeoman AL, Jamison BA, Yeung DT, Scott HS, Hughes TP, et al. BCR-ABL1 kinase ___domain mutations may persist at very low levels for many years and lead to subsequent TKI resistance. Br J Cancer. 2013;109:1593–8.
Soverini S, De Benedittis C, Machova Polakova K, Brouckova A, Horner D, Iacono M, et al. Unraveling the complexity of tyrosine kinase inhibitor-resistant populations by ultra-deep sequencing of the BCR-ABL kinase ___domain. Blood. 2013;122:1634–48.
Parker WT, Lawrence RM, Ho M, Irwin DL, Scott HS, Hughes TP, et al. Sensitive detection of BCR-ABL1 mutations in patients with chronic myeloid leukemia after imatinib resistance is predictive of outcome during subsequent therapy. J Clin Oncol. 2011;29:4250–9.
Smith CC, Brown M, Parker WT, Lin K, Travers K, Wang S, et al. Single Molecule Real Time (SMRT (TM)) sequencing sensitively detects the evolution of Polyclonal and Compound BCR-ABL mutations in patients who relapse on kinase inhibitor therapy. Blood. 2012;120:917.
Kastner R, Zopf A, Preuner S, Proll J, Niklas N, Foskett P, et al. Rapid identification of compound mutations in patients with Philadelphia-positive leukaemias by long-range next generation sequencing. Eur J Cancer. 2014;50:793–800.
Olmedillas-Lopez S, Garcia-Arranz M, Garcia-Olmo D. Current and emerging applications of droplet digital PCR in Oncology. Mol Diagn Ther. 2017;21:493–510.
Soverini S, De Santis S, Martelli M, Monaldi C, Castagnetti F, Gugliotta G, et al. Droplet digital PCR for the detection of second-generation tyrosine kinase inhibitor-resistant BCR::ABL1 kinase ___domain mutations in chronic myeloid leukemia. Leukemia. 2022;36:2250–60.
Vannuffel P, Bavaro L, Nollet F, Aynaci A, Martelli M, Devos H, et al. Droplet Digital PCR Phasing (DROP-PHASE): A Novel Method for Straightforward Detection of BCR-ABL1 Compound Mutations in Tyrosine Kinase Inhibitors Resistant Chronic Myeloid Leukemia (CML) and Acute Lymphoblastic Leukemia (ALL). Blood. 2019;134:4660–4660.
Pont-Kingdon G, Lyon E. Direct molecular haplotyping by melting curve analysis of hybridization probes: beta 2-adrenergic receptor haplotypes as an example. Nucleic Acids Res. 2005;33:e89.
Xu B, Lan Y, Luo D, Zheng Y, Ni R, Su G, et al. Highly sensitive detection of PIK3CA mutations by looping-out probes-based melting curve analysis. Biochem Genet. 2023;62:77–94.
Lan Y, Xu B, Xi Y, Luo Y, Guo X, Huang Z, et al. Accurate detection of multiple tumor mutations in formalin-fixed paraffin-embedded tissues by coupling sequence artifacts elimination and mutation enrichment with meltarray. Lab Invest. 2023;104:100300.
Huang Q, Chen D, Du C, Liu Q, Lin S, Liang L, et al. Highly multiplex PCR assays by coupling the 5’-flap endonuclease activity of Taq DNA polymerase and molecular beacon reporters. Proc Natl Acad Sci USA. 2022;119:e2110672119.
Bidshahri R, Attali D, Fakhfakh K, McNeil K, Karsan A, Won JR, et al. Quantitative detection and resolution of BRAF V600 status in colorectal cancer using droplet Digital PCR and a novel wild-type negative assay. J Mol Diagn. 2016;18:190–204.
Decraene C, Silveira AB, Bidard FC, Vallee A, Michel M, Melaabi S, et al. Multiple hotspot mutations scanning by single droplet digital PCR. Clin Chem. 2018;64:317–28.
Regan JF, Kamitaki N, Legler T, Cooper S, Klitgord N, Karlin-Neumann G, et al. A rapid molecular approach for chromosomal phasing. PLoS One. 2015;10:e0118270.
Parker WT, Phillis SR, Yeung DTO, Hughes TP, Scott HS, Branford S. Many BCR-ABL1 compound mutations reported in chronic myeloid leukemia patients may actually be artifacts due to PCR-mediated recombination. Blood. 2014;124:153–5.
Parker WT, Yeung DT, Yeoman AL, Altamura HK, Jamison BA, Field CR, et al. The impact of multiple low-level BCR-ABL1 mutations on response to ponatinib. Blood. 2016;127:1870–80.
Bullinger L, Dohner K, Dohner H. Genomics of acute myeloid leukemia diagnosis and pathways. J Clin Oncol. 2017;35:934–46.
Soverini S, Martelli M, Bavaro L, De Benedittis C, Sica S, Sora F, et al. BCR-ABL1 compound mutants: prevalence, spectrum and correlation with tyrosine kinase inhibitor resistance in a consecutive series of Philadelphia chromosome-positive leukemia patients analyzed by NGS. Leukemia. 2021;35:2102–7.
Eide CA, Zabriskie MS, Savage Stevens SL, Antelope O, Vellore NA, Than H, et al. Combining the allosteric inhibitor asciminib with ponatinib suppresses emergence of and restores efficacy against highly resistant BCR-ABL1 mutants. Cancer Cell. 2019;36:431–443.e435.
Byrgazov K, Lucini CB, Valent P, Hantschel O, Lion T. BCR-ABL1 compound mutants display differential and dose-dependent responses to ponatinib. Haematologica. 2018;103:e10–e12.
Chen J, Wang F, Fang J, Nie D, Zhang Y, Chen X, et al. Dynamic evolution of ponatinib-resistant mutations in BCR-ABL1-positive leukaemias revealed by next-generation sequencing. Br J Haematol. 2020;191:e113–e116.
Acknowledgements
We thank the patients with leukaemia and their families for providing their specimens for the advancement of cancer research.
Funding
This study was supported by the National Natural Science Foundation of China (82271729); the Natural Science Foundation of Xiamen, China (3502Z20227172); the Science and Technology Planning Projects of Liuzhou City (2023YRZ0103); and the Natural Science and Technology Major Project of Xiamen (3502Z20231034).
Author information
Authors and Affiliations
Contributions
QL, QH, and YL conceived and design the project. YL, PH, WH, AZ, and XG performed the experiments. JD and LX collected peripheral blood and/or bone marrow samples from leukaemia patients. YL and JD collected, analysed and interpreted the data. YL, QH and QL drafted the manuscript. QL, QH, JD, and YH accessed and verified the data. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
This study was approved by the Medical Ethics Committee of Union Hospital affiliated to Tongji Medical College of Huazhong University of Science and Technology (UHCT-IEC-SOP-016-03-01) and performed in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lan, Y., Deng, J., He, P. et al. Innovative rapid screening for complex BCR-ABL1 kinase ___domain mutations in TKI-treated leukaemia patients. Br J Cancer (2025). https://doi.org/10.1038/s41416-025-03098-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41416-025-03098-y