Abstract
The significance of 5-methylcytosine (m5C) methylation in human malignancies has become an increasing focus of investigation. Here, we show that m5C regulators including writers, readers and erasers, are predominantly upregulated in urothelial carcinoma of the bladder (UCB) derived from Sun Yat-sen University Cancer Center and The Cancer Genome Atlas cohort. In addition, NOP2/Sun RNA methyltransferase family member 2 (NSUN2) as a methyltransferase and Aly/REF export factor (ALYREF) as a nuclear m5C reader, are frequently coexpressed in UCB. By applying patient-derived organoids model and orthotopic xenograft mice model, we demonstrate that ALYREF enhances proliferation and invasion of UCB cells in an m5C-dependent manner. Integration of tanscriptome-wide RNA bisulphite sequencing (BisSeq), RNA-sequencing (RNA-seq) and RNA Immunoprecipitation (RIP)-seq analysis revealed that ALYREF specifically binds to hypermethylated m5C site in RAB, member RAS oncogene family like 6 (RABL6) and thymidine kinase 1 (TK1) mRNA via its K171 ___domain. ALYREF controls UCB malignancies through promoting hypermethylated RABL6 and TK1 mRNA for splicing and stabilization. Moreover, ALYREF recognizes hypermethylated m5C site of NSUN2, resulting in NSUN2 upregulation in UCB. Clinically, the patients with high coexpression of ALYREF/RABL6/TK1 axis had the poorest overall survival. Our study unveils an m5C dependent cross-regulation between nuclear reader ALYREF and m5C writer NSUN2 in activation of hypermethylated m5C oncogenic RNA through promoting splicing and maintaining stabilization, consequently leading to tumor progression, which provides profound insights into therapeutic strategy for UCB.
Similar content being viewed by others
Introduction
RNA epigenetic modifications, including N6-methyladenosine (m6A) [1], have been widely implicated functioning in various cellular, developmental, and pathological processes, and determine the fate of RNAs [2, 3]. As one of the most common RNA modification, 5-methylcytosine (m5C) has been identified in tRNAs, rRNAs and mRNAs [4,5,6,7] and plays an essential role in RNA metabolism [8]. The recent research [9] showed that metastasis-initiating tumor cells require mitochondrial m5C to activate invasion and dissemination. mRNA m5C methylation was initially catalyzed by NOP2/Sun RNA methyltransferase family member 2 (NSUN2) and enriched in the vicinity of translational start codon and 3′ untranslated region (UTR) [10, 11]. Huang et al. [12] reported an improved method to identify mRNA m5C sites and determined sequence motifs. Li et al. [13] stratified m5C sites to two types: type I m5C sites contained a downstream G-rich triplet motif; type II m5C sites contain a downstream UCCA motif. Aly/REF export factor (ALYREF) has been identified as the first nuclear m5C reader [14]. Y-box protein 1 (YBX1) [15, 16] has been characterized as the first cytoplasmic m5C reader, maintaining the stability of its targeted m5C transcripts.
So far, it has been evidenced that m6A RNA methylation played an important role in cancer occurrence and development [1, 17,18,19]. The significance of m5C methylation in human malignancies has become an increasing focus of investigation. It was reported that activation of RNA m5C modification was critical for tumor-initiating cells fate and global protein synthesis [20]. We have previously revealed that m5C is preferentially hypermethylated in urothelial carcinoma of the bladder (UCB) and represents a novel mechanism for oncogene activation [21]. As the governor of m5C methylation, m5C regulators, including writers, readers and erasers, play central roles in tumor pathogenesis. Dysregulated expressed m5C regulators in human cancers have been reported in several studies [22,23,24]. We previously reported that cytoplasmic m5C reader YBX1 stabilizes HDGF mRNA, leading to enhanced tumor progression in UCB [21]. m5C writer NSUN2 acts as an oncogene to promote gastric cancer [25] and hepatocellular carcinoma (HCC) [26] development by regulating protein-encoding gene CDKN1C and lncRNA H19, respectively. Wang et al. [27] reported that m5C-methylated PKM2 mRNA was recognized by ALYREF and promoted the glucose metabolism in UCB. Despite these studies, more detailed investigations focusing on the collaboration network among these m5C regulators are still lacking.
UCB is one of the most malignant cancers [28], with a recurrence rate of up to 74% among non-muscle-invasive bladder cancer patients [29]. For muscle-invasive bladder cancer, up to 50% of patients die from distant metastases despite undergoing radical cystectomy with pelvic lymph node dissection [30]. It was reported that 70%-80% bladder cancer patients occurred mutations in the promoter of the gene encoding telomerase reverse transcriptase [31]. Deletions in chromosome 9 [32], mutations in FGFR3 [33] and PI3K [34] were seen as early oncogenic events in UCB. In addition, epigenetic dysregulation may also contribute to the progression of bladder cancer [35]. Our previous studies unveil a novel regulatory mechanism of oncogene activation mediated by m5C methylation in UCB. It is critical to further identify genome-wide m5C methylated genes that function in UCB tumorigenesis.
In this study, we demonstrate that m5C regulators are predominantly upregulated in UCBs from Sun Yat-sen University Cancer Center (SYSUCC) and The Cancer Genome Atlas (TCGA) cohort, and m5C writer NSUN2 and nuclear m5C reader ALYREF are frequently coexpressed. Patient-derived organoids model and orthotopic xenograft mice model showed that ALYREF promotes proliferation and invasion of UCB cells in an m5C-dependent manner. Integration of transcriptome-wide RNA-bisulphite sequencing (BisSeq), RNA-sequencing (RNA-seq) and RNA Immunoprecipitation (RIP)-seq analysis revealed that ALYREF specifically binds to hypermethylated m5C site in RAB, member RAS oncogene family like 6 (RABL6) and thymidine kinase 1 (TK1) mRNA via its K171 ___domain. Mechanistically, ALYREF controls UCB malignancies through promoting hypermethylated RABL6 and TK1 mRNA for splicing and maintaining stabilization. Moreover, ALYREF recognizes hypermethylated m5C site of NSUN2, resulting in NSUN2 upregulation in UCB. Clinically, triple expression of high levels of ALYREF/RABL6/TK1 predict the poorest survival.
Results
m5C regulators are predominantly upregulated in UCB
To investigate the roles of m5C regulators in UCB malignancy, we analyzed the expression profile of m5C regulators in UCB derived from SYSUCC and TCGA cohort. By analyzing our previously published RNA-seq data of 22 paired normal and adjacent UCB tumor tissues, we identified that 7 writers (NOP2, NSUN2, NSUN3, NSUN4, NSUN5, NSUN6 and NSUN7), one reader (ALYREF) and two erasers (TET2 and TET3) were statistically significant and recurrent upregulated in UCB tumor tissues (fold change > 1.1, P-value <0.05, occurrence rate> 50%) (Fig. 1A). Moreover, five writers (NOP2, NSUN2, NSUN3, NSUN4 and NSUN5), one reader (ALYREF) and two erasers (TET2 and TET3) were consistently upregulated in UCB tumor tissues compared to adjacent normal tissues from TCGA cohort (fold change > 1.1, P-value <0.05, occurrence rate > 50%) (Fig. 1B). Among these two cohorts, we found that the expression of ALYREF and NSUN2 were significantly upregulated in UCB tumor tissues compared to adjacent normal tissues (Fig. 1C, D). The expression pattern of other m5C regulators were shown in Fig. S1A and S1B.
A, B Heat maps of m5C regulator mRNA expression profiles in paired UCB tumor tissues and adjacent normal tissues (22 pairs from SYSUCC cohort [A] and 19 pairs from TCGA cohort [B]). C, D The expression levels of ALYREF and NSUN2 (fold change > 1.1, P <0.05) in the SYSUCC cohort (22 pairs) (C) and TCGA cohort (19 pairs) (D). Data represent the mean ± S.D., and a two-tailed paired Student’s t-test was applied to determine the P-value. E The signal pathways in which m5C regulators may be involved from RNA-seq on SYSUCC UCB cohort. Positive correlations are shown in red; negative correlations are shown in blue. F WGCNA showing the networks among m5C regulators in the hierarchical cluster tree.
To investigate the potential function of these m5C regulators in UCB, we analyzed the signal pathways in which m5C regulators may be involved from RNA-seq on SYSUCC UCB cohort. The expression levels of m5C regulators were positively associated with multiple oncogenic pathways, such as K-RAS signaling, oxidative phosphorylation, and TGF-β signaling. Meanwhile, tumor suppressor pathway, such as P53 pathway was negatively associated with m5C regulator expression (Fig. 1E). These results together suggest the essential role of m5C regulators in cancer progression.
To explore the networks among m5C regulators, we conducted Weighted Gene Coexpression Network Analysis (WGCNA) on the UCB TCGA cohort. Notably, m5C writers (NSUN2, NSUN3 and NSUN5) and erasers (TET1, TET2 and TET3) were coexpressed with nuclear reader ALYREF in an mRNA module (Fig. 1F). Our finding indicates that cross-talks among writers, erasers and readers may exist in the m5C regulation, in which ALYREF serves as a core factor, and some m5C regulators may function synergistically. Collectively, these results strongly support that m5C regulators may link to UCB pathogenesis.
Next, we explored the potential function of these m5C regulators candidates in UCB cells. NSUN3, NSUN5, TET2 and TET3 were knocked down by siRNAs in T24 cells (Fig. S1C). Colony-formation and migration assays demonstrated that these four m5C regulators have no significant roles in the proliferation and migration of UCB cells (Fig. S1D and E).
ALYREF is upregulated in UCB and correlates with poor overall survival (OS) in UCB patients
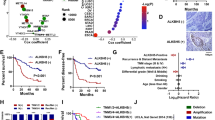
We investigated the clinical significance of ALYREF expression in UCB. Western blotting assay of samples from 10 UCB patients from SYSUCC showed that ALYREF was frequently upregulated in UCB tissues (Fig. 2A). Immunohistochemistry (IHC) staining was performed in a cohort of 170 UCB tissues and 30 paired nonneoplastic bladder tissues from SYSUCC. High ALYREF expression was observed in 99/170 (58.2%) UCB patients. The representative IHC staining images in nonneoplastic bladder tissues and UCB tissues were shown in Fig. 2B. High expression of ALYREF was associated with poor OS in UCB patients significantly (Fig. 2C). The http://gepia2.cancer-pku.cn/#index., which analyzed TCGA cohort, showed that patients with high mRNA level of ALYREF predicted poorer OS (Fig. 2D). These data provide evidence that ALYREF is a potential oncogene in human UCB.
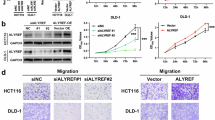
A Western blotting showing ALYREF expression in 10 pairs of UCB and adjacent non-neoplastic tissues. The expression was normalized by α-tubulin expression. T, tumor tissue; N, nonneoplastic bladder tissues. B IHC staining assays and representative images of ALYREF in nonneoplastic bladder tissues (Left) and UCB tissues (Medium and Right). Scale bars, 100 μm. C Kaplan–Meier analysis showing that upregulated ALYREF predicts poor OS in the SYSUCC cohort. The P-value was calculated by a log-rank test. D Kaplan–Meier analysis showed patients with high mRNA expression level of ALYREF predicted poorer OS. The P-value was calculated by a log-rank test. The group cutoff was 50%, which was the expression threshold for splitting the high-expression and low-expression cohorts. The data were from the public database http://gepia2.cancer-pku.cn/#index. E Organoid model showing the growth effect transfected with shCTRL and shALYREF#3 (Left). Statistical analysis of organoid size after 7 days of infection with shCTRL and shALYREF#3 (Right). Scale bars, 100 μm. Data represent the mean ± S.D., n = 3, and a two-tailed unpaired Student’s t-test was applied to determine the P-value. F Representative Hematoxylin-eosin staining images of organoids after infection with shCTRL (Left) and shALYREF#3 (Right). Scale bars: 100 μm. G Colony forming assay showing the effect of ALYREF with a WT m5C site on the restoration of cell growth in ALYREF-knockdown cells relative to ALYREF with K171A mutant. Left: representative images of cell colonies in T24 (Top) and UM-UC-3 (Bottom) cells; Right: histograms of colony numbers. Data represent the mean ± S.D., n = 3. A two-tailed unpaired Student’s t-test was applied to calculate the P-value. H Migration assay showing the effect of ALYREF with a WT m5C site on the restoration of cell migration in ALYREF-knockdown cells relative to ALYREF with K171A mutant. Left: representative images of migration cells in T24 (Top) and UM-UC-3 (Bottom) cells; Scale bars, 100 μm. Right: histograms of the number of migration cells. Data represent the mean ± S.D., n = 3. A two-tailed unpaired Student’s t-test was applied to calculate the P-value.
ALYREF enhances UCB cell proliferation and invasion in an m5C-dependent manner
Patient-derived organoids serve as an ideal cell model to study tumor pathogenesis [36,37,38]. To further explore the role of ALYREF in UCB aggressiveness, we constructed a patient-derived organoid in vitro. ALYREF was knocked down in T24 and UM-UC-3 cells by two short hairpin RNAs (shRNA-2 and shRNA-3) (Fig. S2A). We found that UCB organoid growth was significantly inhibited after knockdown of ALYREF (Fig. 2E, F). Colony-formation and migration assays demonstrated that cell growth and migration abilities were largely reduced after knockdown of ALYREF (Fig. S2B and S2C). We further found that ALYREF did not affect cell growth and migration abilities in a normal urothelial cell line, SV-HUC-1 (Fig. S2A, S2D and S2E). We then explored if the oncogenic function of ALYREF relies on m5C recognition capacity. It has been reported [14] that ALYREF K171A mutation led to a strongly reduced ALYREF binding ability to m5C-containing oligonucleotide, we therefore examined whether ALYREFY K171A mutation could affect the function of ALYREF. We overexpressed shALYREF#3-insensitive wild-type (WT) or the K171A-mutant ALYREF in ALYREF-depleted UCB cells, respectively. The downregulated expression of ALYREF in ALYREF-depleted UCB cells was rescued by overexpression WT or the K171A-mutant ALYREF (Fig. S2F). We further found that after knockdown of ALYREF, colony-formation and cell counting kit-8 (CCK8) assays showed that the reduced tumor cell growth could be rescued by the overexpression of WT ALYREF, but not the K171A-mutant ALYREF (Fig. 2G and Fig. S2G). Migration and invasion assays showed that the reduced migration and invasion capacity could be rescued by the overexpression of WT ALYREF, but not the K171A-mutant ALYREF (Fig. 2H and Fig. S2H). On the contrary, the cell growth and migration abilities of the cells subjected to ALYREF overexpression were significantly increased compared with those of control cells (Fig. S2A, S2I and S2J). Our data indicate that ALYREF exerts oncogenic effects in UCB cells in an m5C-dependent manner.
An orthotopic xenograft model was used to investigate the role of ALYREF in UCB aggressiveness in vivo. Depletion of ALYREF caused fewer submucosal lesions in mice bladder, and this effect could be rescued by overexpression of WT ALYREF but not K171A-mutant ALYREF (Fig. 3A, B and Fig. S3A and S3B). Tumorigenicity assays in vivo showed that knockdown of ALYREF inhibited subcutaneous tumor formation abilities. However, the reduction of subcutaneous tumor formation abilities could be rescued by the overexpression of WT ALYREF but not the K171A-mutant of ALYREF (Fig. 3C and Fig. S3C). Tail-vein injection metastasis assays in vivo demonstrated that ALYREF depletion inhibited lung metastatic nodules formation. The effect of ALYREF knockdown on tumor cell invasion and lung metastasis was rescued by overexpression of WT ALYREF rather than the mutant (Fig. 3D, E). Together, these results show that ALYREF promotes UCB cell proliferation and invasion in an m5C-dependent manner.
A The orthotopic xenograft model showing the effect of ALYREF with a WT m5C site on the restoration of submucosal lesions in ALYREF-knockdown cells relative to ALYREF with K171A mutant. Left: representative bioluminescence images; Right: statistical results for the bioluminescence signals. Data show the mean ± S.D. The P-values were calculated by a two-tailed unpaired Student’s t-test. n = 5, ns: no significance. B Representative Hematoxylin-eosin staining images in different groups of orthotopic xenograft models. Scale bars, 100 μm. C The subcutaneous xenograft model showing the effect of ALYREF with a WT m5C site on the restoration of subcutaneous tumor formation in ALYREF-knockdown cells relative to ALYREF with K171A mutant. D The lung metastasis model showing the effect of ALYREF with a WT m5C site on the restoration of tumor metastasis in ALYREF-knockdown cells relative to ALYREF with K171A mutant. Left: representative bioluminescence images are shown at 0 and the 6th week after injection; Right: statistical results for the mean bioluminescence signals in different groups at the 6th week. Data show the mean ± S.D. The P-values were calculated by a two-tailed unpaired Student’s t-test. n = 5, ns: no significance. E Left: Hematoxylin-eosin staining and metastatic nodules (indicated by arrows) in lung tissues from different groups at the 6th week. Scale bars: 400 µm; Right: Statistical results for the number of metastatic nodules in the lung among different groups at the 6th week. Data show the mean ± S.D, The P-values were calculated by a two-tailed unpaired Student’s t-test. n = 5, ns: no significance.
ALYREF targets hypermethylated m5C site in RABL6 and TK1 mRNA
To further identify the potential mRNAs regulated by ALYREF, we performed RNA-seq analysis in control or ALYREF knockdown T24 cells. After knockdown of ALYREF, 143 mRNAs differentially expressed, including 94 downregulated mRNAs (NC reads >100, |Fold change | >1.5, P-value <0.05). Functional enrichment analysis using Kyoto Encyclopedia of Genes and Genomes (KEGG) indicated that regulated mRNAs by ALYREF are chiefly enriched in canonical cancer-related pathways (Fig. S4A), including TGF-β signaling, MAPK signaling, and NF-κB signaling, strongly supporting the oncogenic function of ALYREF in tumor progression. Among these 94 downregulated genes, 11 mRNA showed a significant reduction in m5C methylation after NUSN2 silencing in T24 cells, combined with previously transcriptome-wide RNA-BisSeq data of T24 cells [21] (Fig. 4A). We further combined RNA-BisSeq in HeLa cells from Yang et al. [14] and Huang et al. [12], and found 3 mRNAs showed a significant decrease in m5C methylation after NUSN2 silencing (Table S7). We next analyzed the ALYREF-RIP-BisSeq from Yang et al. [14] and identified that m5C methylated RABL6 (chr9: 139702478) and TK1 (chr17: 76170268) were located in ALYREF-RIP RNAs (Fig. 4B) (Table S8).
A Venn diagram showing downregulated mRNAs after ALYREF was knocked down and low methylated mRNAs after NSUN2 silencing in T24 cells. Eleven mRNAs are in the intersection. B A flowchart illustrated the screening strategy of ALYREF/NSUN2 targeted candidate genes through m5C regulation. C Silencing NSUN2 reduced the enrichment of m5C level in RABL6 and TK1. Left: Dot blotting of m5C in siCTRL and siNSUN2 in T24 cells. Right: m5C-RIP-qRT-PCR showing the m5C level of RABL6 and TK1 in siCTRL and siNSUN2 cells. Data represent the mean ± S.D., n = 3. A two-tailed unpaired Student’s t-test was applied to calculate the P-value. D Integrative- genomics-viewer tracks representing the read regions of RABL6 (Top) and TK1 (Bottom) in shALYREF#3 RNA-seq data, the m5C sites when NSUN2 was silenced and the ALYREF-binding regions in the RIP-seq data. The triangle indicates the m5C site in RABL6 (chr9: 139702478) and in TK1 (chr17: 76170268), respectively. E RIP assays showing the association of ALYREF with the m5C sites of RABL6, and TK1 mRNAs. Upper panel: western blotting shows the ALYREF IP efficiency in control and shALYREF#3 cells. Bottom panel: Relative enrichment representing RABL6, and TK1 mRNA levels associated with ALYREF compared to an input control. IgG antibody used as a control. Data show the mean ± S.D., n = 3. The P-values were calculated by a two-tailed unpaired Student’s t-test. F qRT-PCR showing the expression change of RABL6 and TK1 mRNA after ALYREF knockdown. (|log2FC | = 1.125, P <0.0001 for RABL6 and |log2FC | = 1.503, P <0.0001 for TK1). Data show the mean ± S.D., n = 3. The P-values were calculated by a two-tailed unpaired Student’s t-test. G Silver staining assays showing the protein bands binding to endogenous ALYREF. H The splicing efficiency of RABL6 (Left) and TK1 (Right) mRNA were calculated by the ratio of spliced to unspliced transcripts. Schematic illustration showing the qRT-PCR primers designed across exon-intron junction and across exon-exon junction. Data show the mean ± S.D., n = 3. The P-values were calculated by a two-tailed unpaired Student’s t-test. I Cytoplasmic and nuclear mRNA fractionation experiment showing the effect of ALYREF with a WT m5C site on the restoration of increased nuclear RABL6 and TK1 content in ALYREF-knockdown cells relative to ALYREF with K171A mutant. Data show the mean ± S.D., n = 3. The P-values were calculated by a two-tailed unpaired Student’s t-test. J RNA stability assay showing RABL6 (Left) and TK1 (Right) mRNA half-life in T24 cells transfected with WT or K171A mutant plasmids after the knockdown of ALYREF. n = 3. The P-values were calculated by a two-tailed unpaired Student’s t-test. K Western blotting showing the protein expression of ALYREF, RABL6 and TK1 in control and shALYREF#3 T24 cells, which expressing WT ALYREF and K171A mutant ALYREF and were normalized by α-tubulin expression. L Western blotting assays showing the protein expression level of RABL6, TK1 and NSUN2 in siCTRL and siNSUN2 cells and were normalized by α-tubulin expression.
According to the RNA-BisSeq from Chen et al. [21], we found that knockdown of NSUN2, the m5C level of RABL6 (chr9: 139702478) was reduced from 0.3528 to 0.1386, while the m5C level of TK1 (chr17: 76170268) was reduced from 0.164 to 0. For further validation, we performed m5C-RIP- quantitative real-time polymerase chain reaction (qRT-PCR) and found that knockdown of NUSN2 substantially reduced the m5C level of RABL6 and TK1 (Fig. 4C). These results together demonstrated that RABL6 contains m5C site in the 5′UTR (chr9: 139702478); TK1 contains m5C site in the 3′UTR (chr17: 76170268). We next performed RNA immunoprecipitation-sequencing (RIP-seq) and RIP-qRT-PCR to identify ALYREF binding targets. ALYREF-Flag-RIP seq from Yang et al. [14] (Table S9) and our RIP-seq confirmed that ALYREF interacted with the m5C sites of RABL6 and TK1 mRNA (Fig. 4D and S4B). Then, we conducted RIP-qRT-PCR analysis by endogenous ALYREF to confirm the binding to targeted mRNAs. When ALYREF was depleted, the relative enrichment of RABL6 and TK1 mRNA was reduced (Fig. 4E). Through qRT-PCR assay, we found the expression of TK1 and RABL6 mRNA were dramatically reduced by ALYREF depletion (|log2FC | > 1, P <0.0001) (Fig. 4F). These results suggest RABL6 and TK1 are direct targets of NSUN2 and ALYREF mediated m5C methylation or recognition.
ALYREF promotes RABL6 and TK1 splicing and maintains their stabilization
To unveil the biological significance of m5C methylation through ALYREF recognition, we purified ALYREF-bound proteins subjected to mass spectrometry analysis (Fig. 4G and S4C). The result showed that several spliceosome factors bound to ALYREF (Fig. S4D), such as SRSF3, PRPF3 and DHX16, indicating ALYREF may function in the regulation of mRNA splicing. We applied iREAD (intron REtention Analysis and Detector) [39] to analyze the reads of shCTRL and shALYREF#3 RNA-seq and found intron retention events in RABL6 and TK1 after ALYREF knockdown (Fig. S4E). We therefore investigated whether ALYREF affects the splicing of RABL6 and TK1 mRNA. The splicing efficiency was determined by qRT-PCR, whereas exon-intron pair amplifies premature isoform mRNA, exon-exon pair amplifies mature form mRNA. After knockdown of ALYREF, the splicing efficiency of RABL6 and TK1 was significantly decreased as measured by the ratio of spliced/ unspliced intermediates. Moreover, exogenous expression of WT ALYREF, but not the K171A mutant of ALYREF, restored the splicing efficiency of RABL6 and TK1 (Fig. 4H). Further analysis showed ALYREF knockdown reduced the level of mature RABL6 and TK1 mRNA, but did not affect the level of premature RABL6 and TK1(Fig. S4F). Similarly, we found that NSUN2 knockdown did not affect the level of premature RABL6 and TK1. However, the level of mature RABL6 and TK1 mRNA was downregulated in NSUN2 knockdown cells (Fig. S4G). As the improperly spliced mRNAs are retained in the nucleus for RNA quality check [40], therefore we determined whether ALYREF recognition of m5C methylated mRNA facilitated mRNA export. We isolated nuclear and cytoplasmic RNA fractions and quantified the quantity of RABL6 and TK1 in each fraction by qRT-PCR. We found that after depletion of ALYREF, RABL6 and TK1 mRNAs was retained in the nucleus. Overexpression of exogenous WT ALYREF, but not the K171A mutant of ALYREF, restored the proper export of RABL6 and TK1 mRNA (Fig. 4I). We further investigated the RNA stability of RABL6 and TK1 by ALYREF depletion. After treatment with actinomycin D, the stability of RABL6 and TK1 was strongly decreased by depletion of ALYREF, while this reduction could be rescued by exogenous WT ALYREF, but not the K171A mutant ALYREF (Fig. 4J). Luciferase reporter assays showed that ALYREF depletion substantially reduced the luciferase mRNA expression and activity of RABL6 with WT m5C site (RABL6-WT) and TK1 with WT m5C site (TK1-WT), but not RABL6 with mutant m5C site (RABL6-Mut) and TK1 with mutant m5C site (TK1-Mut) (Fig. S4H and S4I). In accordance with these results, RABL6 and TK1 protein expression were strongly diminished after ALYREF or NSUN2 depletion (Fig. 4K, L). Overexpression of exogenous WT ALYREF, but not the K171A mutant of ALYREF, restored RABL6 and TK1 protein expression (Fig. 4K). These results suggest that m5C methylation through ALYREF recognition facilitates splicing and maintain stabilization, which consequently leads to proper mRNA export and protein expression.
To further explore whether the K171A mutation impairs the binding of ALYREF to RNA in general, we performed RIP-qRT-PCR analysis to determine the general binding ability of ALYREF K171A mutant to RNA. We found that ALYREF WT binds to m5C sites of RABL6, while ALYREF K171A mutant showed lower level of binding ability to m5C sites of RABL6. Moreover, ALYREF WT and K171A mutants showed the similar binding ability to RBM26 (chr13:79893003-79980390), SLC39A9 (chr14:69865409-69929107), and NUMB (chr14:73741918-73925286), which don’t contain m5C sites from studies of Yang et al. [14], Huang et al. [12] and Chen et al. [21] (Fig. S4J). These data indicate that K171A mutation does not affect general binding ability of ALYREF to RNA.
ALYREF enhances UCB pathogenesis in an m5C-dependent manner
To further determine the pathological significance of m5C methylation at RABL6 and TK1 mRNA, we analyzed previous RNA-BisSeq from SYSUCC cohort. The result indicated that the m5C level of RABL6 was higher in tumor tissue than that in normal tissues (Fig. 5A). We then collected 5 pairs of UCB and normal tissues and extracted RNA to conduct m5C-RIP-qRT-PCR. The results showed an m5C hypermethylation of RABL6 and TK1 in tumors compared to the normal tissues (Fig. 5B). These results indicated potential oncogenic roles of RABL6 and TK1 m5C methylation in UCB progression. We next constructed siRNA-insensitive RABL6 and TK1 expression plasmids either with WT m5C-site (WT Ins) or mutated m5C-site (Mut Ins) to investigate the pathological significance of m5C methylation at RABL6 and TK1 mRNA (Fig. S5A–D). We next explored whether the mutant could affect the m5C level of RABL6 and TK1. We conducted m5C-RIP-qRT-PCR in T24 cells transferred RABL6-WT, RABL6-Mut, TK1-WT and TK1-Mut, respectively. As showed in Fig. S5E, the relative enrichment of m5C level was reduced significantly in RABL6-Mut and TK1-Mut cells compared with RABL6-WT and TK1-Mut, respectively. The colony-formation assay showed that knockdown of RABL6 or TK1 could significantly reduce colony-formation ability, and this reduction could be recovered by RABL6 or TK1 with WT m5C-site but not by m5C site- mutated RABL6 or TK1 (Figs. 5C, D and S5F, G). These findings suggest hypermethylated RABL6 and TK1 promote UCB pathogenesis.
A The m5C level of RABL6 mRNA in 36 UCBs and in 29 adjacent non-neoplastic tissues from SYSUCC. Data represent the mean ± S.D. The P-values were calculated by a two-tailed unpaired Student’s t-test. B m5C-RIP-qRT-PCR showing the relative enrichment of m5C level of RABL6 and TK1 was upregulated in UCB. Left: m5C-RIP-qRT-PCR showing the relative enrichment of m5C level of RABL6 in 5 pairs of UCB and normal tissues. Right: m5C-RIP-qRT-PCR showing the relative enrichment of m5C level of TK1 in 5 pairs of UCB and normal tissues. Data represent the mean ± S.D., n = 5, and a two-tailed paired Student’s t-test was applied to determine the P-value. C Colony forming assay showing the effect of RABL6 with a WT m5C site on the restoration of cell growth in RABL6-knockdown cells relative to RABL6 with a mutated m5C site. Top: representative images of cell colonies; Bottom: histograms of colony numbers. Data show the mean ± S.D., n = 3. The P-values were calculated by a two-tailed unpaired Student’s t-test. D Colony forming assay showing the effect of TK1 with a WT m5C site on the restoration of cell growth in TK1-knockdown cells relative to TK1 with a mutated m5C site. Top: representative images of cell colonies; Bottom: histograms of colony numbers. Data show the mean ± S.D., n = 3. The P-values were calculated by a two-tailed unpaired Student’s t-test. E Colony forming assay showing the effect of RABL6 with a WT m5C site on the restoration of cell growth in ALYREF-knockdown cells. Top: representative images of cell colonies; Bottom: histograms of colony numbers. Data show the mean ± S.D., n = 3. The P-values were calculated by a two-tailed unpaired Student’s t-test. F Left: The subcutaneous xenograft model showing the effect of RABL6 with a WT m5C site on the restoration of subcutaneous tumor formation in ALYREF-knockdown cells. Right: statistical results for the mean tumor weight in different groups. Data show the mean ± S.D. The P-values were calculated by a two-tailed unpaired Student’s t-test. n = 5. G Colony forming assay showing the effect of TK1 with a WT m5C site on the restoration of cell growth in ALYREF-knockdown cells. Top: representative images of cell colonies; Bottom: histograms of colony numbers. Data show the mean ± S.D., n = 3. The P-values were calculated by a two-tailed unpaired Student’s t-test. H Left: The subcutaneous xenograft model showing the effect of TK1 with a WT m5C site on the restoration of subcutaneous tumor formation in ALYREF-knockdown cells. Right: Statistical results for the mean tumor weight in different groups of the subcutaneous xenograft model. Data indicates the mean ± S.D. The P-values were calculated by a two-tailed unpaired Student’s t-test. n = 5. I Migration assay showing the effect of TK1 with a WT m5C site on the restoration of cell migration in ALYREF-knockdown cells. Top: representative images of migration cells; Bottom: histograms of migration cell numbers. Data show the mean ± S.D., n = 3. The P-values were calculated by a two-tailed unpaired Student’s t-test. J The lung metastasis model showing the effect of TK1 with a WT m5C site on the restoration of tumor metastasis in ALYREF-knockdown cells. Representative bioluminescence images are shown at 0 and the 6th week after injection. K Hematoxylin-eosin staining and metastatic nodules (indicated by arrows) in lung tissues from different groups at the 6th week. Scale bars: 100 µm.
To further investigate the functional correlation between ALYREF and RABL6 and TK1, we conducted rescue experiments respectively. We overexpressed RABL6 with WT m5C-site in ALYREF-knockdown T24 cells (Fig. S5H). After knockdown of ALYREF, the inhibited colony formation and subcutaneous tumor formation were partially rescued by expressing RABL6 with WT m5C-site (Fig. 5E and F). Migration assays showed that RABL6 could not rescued reduced tumor cell migration capacity caused by ALYREF (Fig. S5I). Next, we overexpressed TK1 with WT m5C-site in ALYREF-knockdown T24 cells (Fig. S5J). After knockdown of ALYREF, the inhibited colony formation and subcutaneous tumor formation were partially rescued by expressing TK1 with WT m5C-site (Fig. 5G and H). Migration assays and tail-vein injection metastasis assays showed that the inhibited cell migration and lung metastasis were partially rescued by expressing TK1 with WT m5C-site (Fig. 5I–K and Fig. S5K and S5L).
Taken together, these findings demonstrate that ALYREF promote UCB pathogenesis in an m5C-dependent manner.
ALYREF recognizes hypermethylated m5C site of NSUN2, resulting in NSUN2 upregulation in UCB
From our previous RNA-BisSeq data in T24 cell, we identified an m5C methylation site located in the 3′UTR of NSUN2 (chr5: 6600023). Knockdown of NSUN2 significantly reduced the m5C methylation level of NSUN2 (Fig. 6A). This m5C site was accordance with the result from the RNA-BisSeq data from Huang et al. [12] and RNA-BisSeq data from Yang et al. [14] (Table S7). RNA-BisSeq derived from SYSUCC cohort showed that the m5C methylation level of NSUN2 in UCB tumors was higher than that in normal tissues (Fig. 6B). Additionally, the mRNA expression of NSUN2 was positively associated with the m5C level of NSUN2 mRNA in 36 UCB tissues (Fig. 6C), indicating that NSUN2 expression may be regulated by its mRNA m5C methylation level. To test this hypothesis, we constructed luciferase reporter carried NSUN2 with WT m5C site or NSUN2 with mutated m5C site. As expected, the luciferase mRNA and activity level of NSUN2 containing WT m5C site plasmid was significantly higher compared to NSUN2 containing mutated m5C site plasmid, suggesting that NSUN2 expression requires m5C methylation at its 3′UTR. (Fig. 6D). We then conducted m5C-RIP-qRT-PCR to analyze the relative enrichment of m5C level in NSUN2 mRNA with a WT or mutant m5C-site. The relative enrichment of m5C level was reduced significantly in NSUN2-Mut cells compared with NSUN2-WT (Fig. 6E). To further identify the reader of m5C methylation at NSUN2, we found that the ALYREF-RIP-BisSeq data from Yang et al. [14] showed that m5C methylated NSUN2 (chr5: 6600023) were located in ALYREF-RIP RNAs (Table S8). Our ALYREF RIP-seq data (Fig. 6F) and ALYREF RIP-seq data of Yang et al. [14] (Table S9) showed that the binding site of ALYREF on NSUN2 mRNA coincided well with the m5C site of NSUN2. The specific binding was further confirmed by RIP-qRT-PCR assay (Fig. 6G). Western blotting assays indicated that NSUN2 expression was reduced when ALYREF was knocked down, and the reduction could be rescued by WT but not K171A-mutant ALYREF (Fig. 6H). In addition, by IHC analysis of mice bladder slices from orthotopic xenograft model, the downregulation of NSUN2 was correlated with ALYREF knockdown. WT, but not K171A mutant ALYREF could restore the expression of NSUN2 (Fig. 6I). These data together indicate that ALYREF recognizes hypermethylated m5C site of NSUN2, resulting in NSUN2 upregulation in UCB. Clinically, RNA-seq analysis of the SYSUCC cohort and TCGA cohort showed that NSUN2 and ALYREF levels were positively correlated in UCB (Fig. 6J), suggesting that NSUN2-ALYREF cross-regulation is a bona fide mechanism in UCB progression, contributing to the homeostatic control of RNA m5C methylation.
A Integrative- genomics-viewer tracks representing the methylated level of m5C sites in NSUN2 when NSUN2 was silenced (the methylated level is 0.213 for siCTRL and empty for siNSUN2, respectively). The triangle represents the m5C site in NSUN2 (chr5: 6600023). B The m5C level of NSUN2 in 36 UCBs and in 29 adjacent normal tissues from SYSUCC. A two-tailed unpaired Student’s t-test was applied to determine the P-value. C Pearson correlation analysis showing the association between NSUN2 mRNA expression and its m5C level in 36 UCBs of SYSUCC. Shaded regions represent the 95% confidence interval. D Luciferase reporter assay showing the luciferase mRNA (Left) and activity (Right) level of NSUN2-wild-type m5C site containing plasmid and NSUN2 mutated m5C site containing plasmid in T24 cells. Data represent the mean ± S.D., n = 3. The P-value was determined by a two-tailed unpaired Student’s t-test. E The relative enrichment of m5C level in wild-type NSUN2 containing m5C-site compared with m5C-site mutant NSUN2. Data represent the mean ± S.D., n = 3, and a two-tailed unpaired Student’s t-test was applied to determine the P-value. F Integrative- genomics-viewer tracks representing the read coverage of NSUN2 in ALYREF-Flag RIP-seq data and the m5C levels of 36 UCB and 29 adjacent non-neoplastic tissues from SYSUCC. The triangle indicates the m5C site (chr5: 6600023) in NSUN2. G Upper panel: Western blotting shows Flag IP efficiency between ALYREF WT and K171A mutant. Bottom panel: Relative enrichment representing NSUN2 mRNA levels associated with ALYREF compared to an input control. IgG antibody used as a control. Data show the mean ± S.D., n = 3. The P-values were calculated by a two-tailed unpaired Student’s t-test. H Western blotting assays showing the expression level of NSUN2 and ALYREF in control and shALYREF#3 T24 cells, which expressing WT ALYREF and the K171A mutant and were normalized by α-tubulin expression. I IHC staining assays of mice bladder slices from orthotopic xenograft models showing the effect of ALYREF with a WT m5C site on the restoration of the ALYREF (Left column) and NSUN2 (Right column) expression in ALYREF-knockdown cells relative to ALYREF with K171A mutant. Scale bars, 100 μm. J Pearson correlation analysis showing the association between NSUN2 and ALYREF mRNA expression in the SYSUCC cohort (Left, n = 36) and TCGA cohort (Right, n = 430). Shaded regions showed the 95% confidence interval.
ALYREF-RABL6-TK1 m5C related axis predicts poorest OS in UCB
Next, we examined expression levels of ALYREF and downstream m5C-methylated proteins (RABL6 and TK1) in UCB tissue samples from SYSUCC and TCGA cohort. From RNA-seq analysis, the expression levels of RABL6 and TK1 mRNA are positively associated with the levels of ALYREF mRNA in SYSUCC and TCGA cohort. (Fig. 7A, B). The IHC analysis and double immunofluorescence staining showed that the expression levels of RABL6 and TK1 were positively associated with that of ALYREF in UCBs (Fig. 7C and Fig. S5M). Furthermore, subgroup of individuals with UCBs was classified to investigate the relationship between ALYREF-m5C-related proteins (RABL6 and TK1) and survival rate. Notably, high levels of ALYREF and high levels of RABL6 or TK1 were significantly associated with poorer OS. Triple high expression of ALYREF, RABL6 and TK1 was correlated with the poorest OS in SYSUCC cohort (Fig. 7D). Collectively, these data suggest that ALYREF- RABL6-TK1 m5C-related axis is involved in UCB aggressiveness (Fig. 7E), highlighting its potential as a diagnostic marker and therapeutic target for UCBs.
A Pearson correlation analysis showing the association between RABL6, TK1 and ALYREF mRNA expression in the SYSUCC cohort. n = 36. Shaded regions showed the 95% confidence interval. B Pearson correlation analysis showing the association between RABL6, TK1 and ALYREF mRNA expression in TCGA cohort. n = 430. Shaded regions showed the 95% confidence interval. C Representative IHC staining and double immunofluorescence staining images of ALYREF, RABL6 and TK1 in two UCB tissues with high (the first row) or low expression (the second row). Blue (DAPI) = cell nuclei, red (Cyanine 3) = ALYREF, green (Alexa 488) = RABL6/TK1. Scale bars, 100 μm. D Kaplan–Meier analysis of data of 170 UCB patients from SYSUCC showing the correlation between different expression patterns of ALYREF/ RABL6 (Top, P <0.001) and OS, the correlation between different expression patterns of ALYRREF/TK1 (Medium, P <0.001) and OS, and the correlation between different expression patterns of ALYREF/ RABL6/ TK1 (Bottom, P <0.001), and OS. The P-values were calculated by a log-rank test. E Schematic illustration showing that m5C dependent cross-regulation between nuclear reader ALYREF and writer NSUN2 promotes urothelial bladder cancer malignancy through facilitating mRNAs splicing and stabilization.
Discussion
Epigenetic modifications play essential roles in gene regulation, environmental interactions and cancers [41]. m6A modification has been identified as an important factor in the determination of mammalian cell fate transition, embryonic stem cell differentiation and tumorigenesis [42]. Several studies have suggested that m6A regulators were upregulated in cancers and m6A modification promotes the development of tumors [43,44,45]. Since RNA modifications were controlled by regulators, abnormal expression of these regulators may cause tumorigenesis or cancer progression. As a kind of RNA modifications, m5C plays an important role in cancer tumorigenesis. In this study, by integration of RNA-seq data from SYSUCC and TCGA, we found that m5C regulators including ALYREF are consistently upregulated in UCB compared to normal tissues, and upregulated ALYREF is positively associated with UCB patients’ poorer OS. m5C regulators were positively associated with multiple oncogenic pathways. These results supported that ALYREF may play an essential part in bladder cancer.
We applied the model of patient-derived organoids to explore the function of ALYREF. Several studies demonstrated that organoid models maintain key features from their parental tumors, such as genetic and phenotypic heterogeneity, allowing them to be used for a wide spectrum of applications [36, 38]. In addition, organoids can be established and expanded with high efficiency from primary patient material [46]. Moreover, organoid models showed improved resemblance to the original tumor compared to 2D cultured cancer cell lines [37, 38]. Therefore, we demonstrated that patient-derived organoids serve as an ideal cell model to study tumor pathogenesis. Considering organoid cultures bridge the gap between in vitro 2D cancer cell line cultures and in vivo parental tumors, we thus applied organoids a promising tool to further explored the biological function of ALYREF.
Combined with RNA BisSeq data of Yang et al. [14], Huang et al. [12] and our previous reports, we confirmed RABL6 and TK1 are direct targets of NSUN2. The ALYREF-RIP-BisSeq data from Yang et al. [14] identified m5C methylated RABL6, TK1 and NSUN2 were enriched in ALYREF-RIP RNAs. The ALYREF-RIP-seq from Yang et al. [14] showed ALYREF interacted with m5C methylation sites of RABL6, TK1 and NSUN2. These results suggested that the m5C sites of RABL6, TK1 and NSUN2 were true recognized by ALYREF and regulated by NSUN2 methylation. It has been reported that NSUN2 and NSUN6 play important roles in Type I or Type II-modified m5C in mRNAs [7, 12, 13]. To further analyze RNA secondary structure of m5C site of TK1, RABL6 and NSUN2, we extracted the upstream and downstream 25 bp sequences of the m5C sites (Table S10) and used RNAfold tool (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) to complete this prediction. The analysis showed that the m5C site of TK1 containing a downstream G-rich triplet motif, which may be similar to Type I modified m5C described by Huang et al. [14]. RNAfold prediction revealed TK1 m5C site have a tRNA-like structure. However, the m5C sites of RABL6 and NSUN2 did not contain a downstream G-rich triplet motif, and did not show a tRNA-like structure. Therefore, based on these results and our findings, we demonstrated that m5C sites, which represent tRNA-like structures or tRNA-unlike structures, were both regulated by NSUN2.
RABL6 and TK1 are well-known oncogenes and promote tumor proliferation in many types of cancers [47,48,49,50]. Xu et al. [51] found that circTMC5 sponged miR-361-3p to up-regulate RABL6 expression to promotes gastric cancer. Gandhi et al. [52] showed lincNMR-YBX1 axis regulated TK1 expression by binding its promoter regions. In our study, the mutant of m5C site at RABL6 and TK1 could reduce proliferative capacity of tumors. It is as well reported that the m5C at a particular mRNA position may affect tumor stage. Sun et al. [26] found NSUN2-mediated m5C modification of H19 lncRNA is associated with poor differentiation of HCC. By applying bisulfite-PCR pyrosequencing, they found the methylation level at the H19 C986 site in HCC tissues was significantly higher than that in matched non-cancerous liver tissues. The m5C methylation level of H19 RNA in HCC patients are significantly associated with the differentiation stages of tumors (P <0.001). Our results propose a novel m5C-modification-dependent mechanism of RABL6 and TK1 expression, which contributes in UCB progression.
The removal of introns by splicing is an important step of precursor mRNA process, which frequently altered in tumors [53]. Splicing abnormalities can result in tumor proliferation [54], progression and invasion [55]. Epigenetic modifications including m6A modification, participated in mRNA splicing to regulate tumorigenesis and development. m6A writers like METTL16 [56] and METTL13 [57], m6A readers like YTHDC1 [58], HNRNPA2B1 [59], and m6A erasers like FTO [60] were reported to mediate mRNA splicing to control tumors. Specifically, splicing factors like SRSF3, which interacted with YTHCD1 to promote mRNA splicing and nucleus export of m6A-modified mRNAs, was also found binding to ALYREF in our study and from Khan et al. [61] (Table S11). In addition, TREX complex have been found to bind with endogenous ALYREF from our study and from Khan et al. [61] (Table S11). Similarly, Mendel et al. [62] found that m6A modification was deposited on the 3′ splice site of the S-adenosylmethionine synthetase pre-mRNA, which inhibited proper splicing and protein production. We firstly reported that m5C reader ALYREF promoted UCB malignancy through regulating mRNA splicing via recruiting spliceosome to targeted hypermethylated mRNAs.
Little has been known about the cross-regulations among mRNA methylation regulators. Several studies showed cross-regulation between m6A regulators. Liu et al. [63] demonstrated that the expression of m6A writers was positively correlated with their m6A variation; additionally, conserved m6A peaks of m6A regulators were observed in all human tissues, suggesting that the transcripts of the m6A modification machineries are also susceptible to epitranscriptomic regulation. Panneerdoss et al. [64] revealed that the collaboration among METTL14-ALKBH5-YTHDF3 (writer-eraser-reader) sets up the m6A threshold to regulate the stability of target proliferation-specific gene, resulting in tumor progression. In the current study, we firstly demonstrated that ALYREF recognizes hypermethylated m5C site of NSUN2, resulting in NSUN2 upregulation in UCB. Integration of RNA-BisSeq and RNA-seq in UCB cell and tumor samples, we found that the m5C level of NSUN2 mRNA was positively associated with NSUN2 mRNA expression in SYSUCC cohort, suggesting that NSUN2 expression is regulated by its mRNA m5C methylation level. RIP-seq demonstrated that ALYREF recognizes hypermethylated m5C site of NSUN2, resulting in NSUN2 upregulation in UCB. Together, our study revealed that NSUN2-ALYREF cross-regulation is a bona fide mechanism in UCB progression, contributing to the homeostatic control of RNA m5C methylation.
In summary, our study underlines the significance of m5C methylation in human UCB. We demonstrate that ALYREF enhances proliferation and invasion of UCB cells in an m5C-dependent manner. ALYREF controls UCB malignancies through promoting hypermethylated RABL6 and TK1 mRNA for splicing and stabilization. Moreover, ALYREF recognizes hypermethylated m5C site of NSUN2, resulting in NSUN2 upregulation in UCB. Clinically, triple high expression of ALYREF/RABL6/TK1 axis predicts the poorest survival. Our study unveils a novel m5C dependent cross-regulation between nuclear reader ALYREF and m5C writer NSUN2 in activation of hypermethylated m5C oncogenic RNA, which consequently leads to tumor progression. These findings provide profound insights into therapeutic strategy for the disease.
Materials and methods
Patients and tissue sample collection
Protein samples collected from UCBs and adjacent non-neoplastic tissues of 10 patients who underwent radical cystectomy at SYSUCC were applied for western blotting analyses (Table S1).
A total of 170 UCBs and 30 adjacent non-neoplastic tissues from 170 UCB cases who underwent radical cystectomy from 2005 to 2016 at SYSUCC were used in the IHC analyses (Table S2). The TNM classification and tumor grades were defined in accordance with the eighth edition of the Union for International Cancer Control and the World Health Organization, respectively. Patients were followed up regularly depending on the guidelines. OS was defined as the time from treatment to the date of death due to any cause. After formalin fixation, all samples from these patients were subjected to paraffin-embedding and pathological diagnosis.
For the organoid model, the UCB tissue was collected from a UCB patient who underwent radical cystectomy and had a pathological diagnosis of UCB from SYSUCC (Table S3).
For the m5C-RIP-qRT-PCR, the 5 pairs of UCB and normal tissues were collected from patients receiving radical cystectomy and had a pathological diagnosis of UCB from SYSUCC (Table S4).
Cell cultures
The cell lines used in our study, including SV-HUC-1, T24, UM-UC-3, TCC-SUP, 293 T cell lines, were obtained from American Type Culture Collection. RPMI-1640 medium (Invitrogen, Carlsbad, USA) containing 10% fetal bovine serum (HyClone, USA) was used to culture T24 cells. Other cell types were maintained in DMEM (Invitrogen, Carlsbad, USA) with 10% fetal bovine serum. A humidified incubator at 37 °C with 5% CO2 was provided for culturing cells. Cell lines were authenticated by short tandem repeat profiling and were tested free of mycoplasma contamination using PCR with TaKaRa PCR Mycoplasma Detection Set. All cell lines were cultured within 10 passages.
Western blotting
Extracted proteins were dissolved in 1× SDS and then resolved by SDS-PAGE. After transfer to a PVDF membrane (Millipore, Massachusetts, USA), the membrane was incubated at 4 °C overnight with primary antibodies and room temperature for 1 h with secondary antibodies. The signals on the membranes were showed by an enhanced chemiluminescence kit (Tanon, Shanghai, China). The primary antibodies used for western blotting in our study were as follows: rabbit polyclonal anti-NSUN3 (Abclonal, Cat#: A12892; 1:1000), rabbit polyclonal anti-NSUN5 (Proteintech, Cat#:15449-1-AP; 1:1000), rabbit polyclonal anti-TET2 (Proteintech, Cat#: 21207-1-AP; 1:1000), rabbit polyclonal anti-TET3 (Abclonal, Cat#: A18319; 1:1000), rabbit polyclonal anti-ALYREF (Cell Signaling Technology, Cat#: 12655; 1:1000), rabbit polyclonal anti-NSUN2 (Proteintech, Cat#: 20854-I-AP; 1:5000), rabbit polyclonal anti-Flag-HRP (Cell Signaling Technology, Cat#: 2368 S; 1:1000), anti-α-tubulin (Beyotime, Cat#: AF0001; 1:1000), anti-TK1 (Proteintech, Cat#: 67787-1-Ig; 1:2000), and anti-RABL6 (Proteintech, Cat#: 20848-1-AP; 1:500).
Immunohistochemistry
The obtained organs and tumors were formalin-fixed and paraffin-embedded. Then, 4-µm thick tissue sections were cut for IHC staining. Sections for IHC analysis were first heated at 65 °C for 2 h, deparaffnized in xylene and hydrated in graded alcohol. Endogenous peroxidase activity was inhibited in 3% hydrogen peroxide. Slides were incubated in Ethylenediaminetetraacetic Acid (EDTA) buffer (pH 8.0) for 5 min to retrieve antigen. After blocking nonspecific binding in 10% normal goat serum, primary antibodies for IHC were added for incubation overnight at 4 °C. Before staining with DAB staining solution and restaining with hematoxylin, the slides were incubated with secondary antibodies for 30 min at 37 °C. Seventy percent ethyl alcohol containing 0.1% hydrochloric acid was used to polarize the slides for 10 s.
Evaluation criteria including staining intensity and the positively stained area were applied for IHC staining. Staining intensity was divided into 0, 1, 2, and 3, which indicated no, weak, moderate and strong staining, respectively. The grades for positively stained cells included 1, 2, 3, and 4, which indicated a positively stained area of <10%, 10%–40%, 40%–70% and >70%, respectively. The immunoreactivity score combining the staining intensity and positively stained area scores was calculated by two independent pathologists who were blinded to the clinicopathological information. The primaries antibodies for IHC used in our study were as follows: rabbit polyclonal anti-ALYREF (Cell Signaling Technology, Cat#: 12655; 1:200), rabbit polyclonal anti-NSUN2 (Proteintech, Cat#: 20854-I-AP; 1:200), mouse monoclonal anti-TK1 (Proteintech, Cat#: 67787-1-Ig; 1:250), mouse polyclonal anti-RABL6 (Abnova, Cat#: H00055684-A01; 1:200).
RNA interference
Short hairpin RNAs (shRNAs) used for ALYREF knockdown were acquired from GeneCopeia (Guangzhou, China), while short interfering RNAs (siRNAs) for NUSN3, NSUN5, TET2, TET3, NSUN2, TK1, and RABL6 knockdown were purchased from RIBOBIO (Guangzhou, China). Table S5 shows the sequences which are targeted for siRNAs and shRNAs.
Further materials and methods were shown in supplementary information.
Statistical analysis
Statistical analysis was conducted with SPSS version 23.0 (IBM Corp., Armonk, NY, USA). Statistics are shown as the means ± SD. For analysis of the SYSUCC cohort, the differential expression of genes between UCB and normal tissues was analyzed by two-sided t-tests and the heatmap presenting the difference was generated by the R- package “Heatmap”. To analyze correlations among genes in the TCGA and SYSUCC cohorts, Spearman’s correlation analysis was applied. The Kaplan–Meier method and the log-rank test were conducted for survival analysis. The statistical significance between experimental groups were determined by two-sided t-tests or two-way ANOVA. The composition ratios were analyzed by the chi-square test. All experiments were independently conducted at least three times with similar results.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files and supplementary figure 6. Additional data associated with this paper may be acquired from the corresponding author on reasonable request. The RNA-Seq data is deposited in the SRA database, and the Bioproject number is PRJNA765965.
References
Huang H, Weng H, Chen J. m6A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–88.
Delaunay S, Frye M. RNA modifications regulating cell fate in cancer. Nat Cell Biol. 2019;21:552–9.
Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell 2017;169:1187–200.
Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-___domain modifications. EMBO Rep. 2008;9:629–35.
Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37:e12.
Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–33.
Selmi T, Hussain S, Dietmann S, Heiß M, Borland K, Flad S, et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 2021;49:1006–22.
Trixl L, Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip Rev RNA. 2019;10:e1510.
Sylvain D, Gloria P, Bohai F, Kevin K, Mikaela B, Agnes H, et al. Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature. 2022. https://doi.org/10.1038/s41586-022-04898-5.
Amort T, Rieder D, Wille A, Khokhlova-Cubberley D, Riml C, Trixl L, et al. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017;18:1.
Chen YS, Yang WL, Zhao YL, Yang YG. Dynamic transcriptomic m5C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA. 2021;12:e1639.
Huang T, Chen W, Liu J, Gu N, Zhang R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat Struct Mol Biol. 2019;26:380–8.
Liu J, Huang T, Zhang Y, Zhao XN, Chen WY, Zhang R. Sequence- and structure-selective mRNA m5C methylation by NSUN6 in animals. Natl Sci Rev. 2021;8:nwaa273.
Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017;27:606–25.
Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL, et al. RNA 5-Methylcytosine facilitates the maternal-to-Zygotic transition by preventing maternal mRNA decay. Mol Cell. 2019;75:1188–202. e11.
Nombela P, Miguel-López B, Blanco S. The role of m6A, m(5)C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol Cancer. 2021;20:18.
Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9:124.
Liu J, Harada BT, He C. Regulation of gene expression by N(6)-methyladenosine in cancer. Trends Cell Biol. 2019;29:487–99.
Han X, Wang M, Zhao YL, Yang Y, Yang YG. RNA methylations in human cancers. Semin Cancer Biol. 2021;75:97–115.
Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, et al. Stem cell function and stress response are controlled by protein synthesis. Nature 2016;534:335–40.
Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978–90.
Yu GP, Bao JH, Zhan M, Wang JY, Li XJ, Gu X, et al. Comprehensive analysis of m5C methylation regulatory genes and tumor microenvironment in prostate cancer. Front Immunol. 2022;13:914577.
Tong XY, Xiang YL, Hu YB, Hu YY, Li H, Wang HL, et al. NSUN2 promotes tumor progression and regulates immune infiltration in nasopharyngeal carcinoma. Front Oncol. 2022;12:788801.
Chen SY, Chen KL, Ding LY, Yu CH, Wu HY, Chou YY, et al. RNA bisulfite sequencing reveals NSUN2-mediated suppression of epithelial differentiation in pancreatic cancer. Oncogene 2022;41:3162–76.
Mei L, Shen C, Miao R, Wang JZ, Cao MD, Zhang YS, et al. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57(Kip2) by an m5C-dependent manner. Cell Death Dis. 2020;11:270.
Sun Z, Xue SL, Zhang MY, Xu H, Hu XM, Chen SH, et al. Aberrant NSUN2-mediated m5C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene 2020;39:6906–19.
Wang JZ, Zhu W, Han J, Yang X, Zhou R, Lu HC, et al. The role of the HIF-1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer. Cancer Commun (Lond). 2021;41:560–75.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363–85.
Lobo N, Mount C, Omar K, Nair R, Thurairaja R, Khan MS. Landmarks in the treatment of muscle-invasive bladder cancer. Nat Rev Urol. 2017;14:565–74.
Leão R, Lee D, Figueiredo A, Hermanns T, Wild P, Komosa M, et al. Combined genetic and epigenetic alterations of the TERT promoter affect clinical and biological behavior of bladder cancer. Int J Cancer. 2019;144:1676–84.
Stoehr R, Zietz S, Burger M, Filbeck T, Denzinger S, Obermann EC, et al. Deletions of chromosomes 9 and 8p in histologically normal urothelium of patients with bladder cancer. Eur Urol. 2005;47:58–63.
van Rhijn BW, Vis AN, van der Kwast TH, Kirkels WJ, Radvanyi F, Ooms EC, et al. Molecular grading of urothelial cell carcinoma with fibroblast growth factor receptor 3 and MIB-1 is superior to pathologic grade for the prediction of clinical outcome. J Clin Oncol. 2003;21:1912–21.
López-Knowles E, Hernández S, Malats N, Kogevinas M, Lloreta J, Carrato A, et al. PIK3CA mutations are an early genetic alteration associated with FGFR3 mutations in superficial papillary bladder tumors. Cancer Res. 2016;66:7401–4.
Tran L, Xiao JF, Agarwal N. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21:104–21.
Lee SH, Hu WH, Matulay JT, Silva MV, Owczarek TB, Kim K, et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 2018;173:515–28.
Sachs N, Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev. 2014;24:68–73.
Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 2018;172:373–86.
Li HD, Funk CR, Price ND. iREAD: a tool for intron retention detection from RNA-seq data. BMC Genomics. 2020;21:128.
Wegener M, Müller-McNicoll M. Nuclear retention of mRNAs—quality control, gene regulation and human disease. Semin Cell Dev Biol. 2018;79:131–42.
Haruehanroengra P, Zheng YY, Zhou Y, Huang Y, Sheng J. RNA modifications and cancer. RNA Biol. 2020;17:1560–75.
Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. The RNA modification landscape in human disease. RNA 2017;23:1754–69.
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110.
Cheng M, Sheng L, Gao Q, Xiong QC, Zhang HJ, Wu MQ, et al. The m6A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene 2019;38:3667–80.
Tao L, Mu X, Chen H, Jin D, Zhang RY, Zhao YY, et al. FTO modifies the m6A level of MALAT and promotes bladder cancer progression. Clin Transl Med. 2021;11:e310.
Bleijs M, van de Wetering M, Clevers H, Drost J. Xenograft and organoid model systems in cancer research. EMBO J. 2019;38:e101654.
Montalbano J, Jin W, Sheikh MS, Huang Y. RBEL1 is a novel gene that encodes a nucleocytoplasmic Ras superfamily GTP-binding protein and is overexpressed in breast cancer. J Biol Chem. 2007;282:37640–9.
Kohlmeyer JL, Kaemmer CA, Umesalma S, Gourronc FA, Klingelhutz AJ, Quelle DE. RABL6A regulates schwann cell senescence in an RB1-dependent manner. Int J Mol Sci. 2021;22:5367.
Zhu X, Shi CY, Peng YP, Yin LD, Tu M, Chen QY, et al. Thymidine kinase 1 silencing retards proliferative activity of pancreatic cancer cell via E2F1-TK1-P21 axis. Cell Prolif. 2018;51:e12428.
Yogev O, Almeida GS, Barker KT, George SL, Kwok C. In vivo modeling of chemoresistant neuroblastoma provides new insights into chemorefractory disease and metastasis. Cancer Res. 2019;79:5382–93.
Xu P, Xu XL, Wu X, Zhang LX, Meng L, Chen ZM, et al. CircTMC5 promotes gastric cancer progression and metastasis by targeting miR-361-3p/RABL6. Gastric Cancer. 2021. https://doi.org/10.1007/s10120-021-01220-6.
Gandhi M, Groß M, Holler JM, Coggins SA, Patil N, Leupold JH, et al. The lncRNA lincNMR regulates nucleotide metabolism via a YBX1 - RRM2 axis in cancer. Nat Commun. 2020;11:3214.
Desterro J, Bak-Gordon P, Carmo-Fonseca M. Targeting mRNA processing as an anticancer strategy. Nat Rev Drug Disco. 2020;19:112–29.
Bechara EG, Sebestyén E, Bernardis I, Eyras E, Valcárcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol cell. 2013;52:720–33.
Tripathi V, Shin JH, Stuelten CH, Zhang YE. TGF-β-induced alternative splicing of TAK1 promotes EMT and drug resistance. Oncogene 2019;38:3185–200.
Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, et al. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–14.
Li FX, Yi Y, Miao YY, Long WY, Long T, Chen SY, et al. N6-Methyladenosine modulates nonsense-mediated mRNA decay in human glioblastoma. Cancer Res. 2019;79:5785–98.
Roundtree IA, Luo GZ, Zhang ZJ, Wang X, Zhou T, Cui YQ, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 2017;6:e31311.
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 2015;162:1299–308.
Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–19.
Khan M, Hou S, Azam S, Lei H. Sequence-dependent recruitment of SRSF1 and SRSF7 to intronless lncRNA NKILA promotes nuclear export via the TREX/TAP pathway. Nucleic Acids Res. 2021;49:6420–36.
Mendel M, Delaney K, Pandey RR, Chen KM, Wenda JM, Vågbø CB, et al. Splice site m6A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell 2021;184:3125–42.
Liu J, Li K, Cai J, Zhang M, Zhang X, Xiong X, et al. Landscape and regulation of m6A and m6Am methylome across human and mouse tissues. Mol cell. 2020;77:426–40. e6.
Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S, et al. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci Adv. 2018;4:eaar8263.
Acknowledgements
We thank Prof. Yun-Gui Yang (Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing, China) for providing ALYREF-WT, ALYREF-K171A plasmids. This work was supported by the National Natural Science Foundation of China (grant No. 81972382) and the Postdoctoral Science Foundation of China (grant No.2022M723607).
Author information
Authors and Affiliations
Contributions
NW, RC and WW performed experiments and wrote the manuscript. MD and ZZ conducted data analysis and manuscript revision. KN, YL and XL participated in some experiments and provided important advices. YY and JW provided samples. BD participated in some experiments. XZ, ZL and FZ designed this study and provide the guidance. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
This study was approved by the Institutional Review Board of SYSUCC and complied with the Declaration of Helsinki. Before the study began, the patients signed informed written consent. All procedures involving animals were approved by the Institutional Animal Care and Use Committee of SYSUCC.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Dr Maurizio Fanciulli
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, N., Chen, Rx., Deng, Mh. et al. m5C-dependent cross-regulation between nuclear reader ALYREF and writer NSUN2 promotes urothelial bladder cancer malignancy through facilitating RABL6/TK1 mRNAs splicing and stabilization. Cell Death Dis 14, 139 (2023). https://doi.org/10.1038/s41419-023-05661-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-023-05661-y
This article is cited by
-
Comprehensive analysis of RNA methylation-related genes to identify molecular cluster for predicting prognosis and immune profiles in bladder cancer
Scientific Reports (2025)
-
FOXA1-dependent NSUN2 facilitates the advancement of prostate cancer by preserving TRIM28 mRNA stability in a m5C-dependent manner
npj Precision Oncology (2025)
-
RNA binding protein ALYREF regulates ferroptosis to facilitate LUAD growth and metastasis via promoting SLC7A11 mRNA stability
Scientific Reports (2025)
-
Epigenetics in the modern era of crop improvements
Science China Life Sciences (2025)
-
NSUN2 relies on ALYREF to regulate Nrf2-mediated oxidative stress and alleviate Dox-induced liver injury
Biology Direct (2024)