Abstract
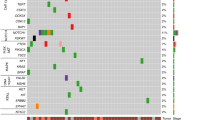
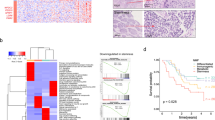
Esophageal squamous cell carcinoma (ESCC) is a poor-prognosis cancer type with limited understanding of its molecular etiology. Using 508 ESCC genomes, we identified five novel significantly mutated genes and uncovered mutational signature clusters associated with metastasis and patients’ outcomes. Several functional assays implicated that NFE2L2 may act as a tumor suppressor in ESCC and that mutations in NFE2L2 probably impaired its tumor-suppressive function, or even conferred oncogenic activities. Additionally, we found that the NFE2L2 mutations were significantly associated with worse prognosis of ESCC. We also identified potential noncoding driver mutations including hotspot mutations in the promoter region of SLC35E2 that were correlated with worse survival. Approximately 5.9% and 15.2% of patients had high tumor mutation burden or actionable mutations, respectively, and may benefit from immunotherapy or targeted therapies. We found clinically relevant coding and noncoding genomic alterations and revealed three major subtypes that robustly predicted patients’ outcomes. Collectively, we report the largest dataset of genomic profiling of ESCC useful for developing ESCC-specific biomarkers for diagnosis and treatment.
Similar content being viewed by others
Login or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Change history
17 February 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41422-022-00625-x
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Chen, W. et al. Cancer statistics in China, 2015. CA Cancer J. Clin. 66, 115–132 (2016).
Lin, Y. et al. Esophageal cancer in high-risk areas of China: research progress and challenges. Ann. Epidemiol. 27, 215–221 (2017).
Lin, D. C., Wang, M. R. & Koeffler, H. P. Genomic and epigenomic aberrations in esophageal squamous cell carcinoma and implications for patients. Gastroenterology 154, 374–389 (2018).
Gao, Y. B. et al. Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 46, 1097–1102 (2014).
Song, Y. et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 509, 91–95 (2014).
Lin, D. C. et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 46, 467–473 (2014).
Zhang, L. et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am. J. Hum. Genet. 96, 597–611 (2015).
Chang, J. et al. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nat. Commun. 8, 15290 (2017).
Cancer Genome Atlas Research, N. et al. Integrated genomic characterization of oesophageal carcinoma. Nature 541, 169–175 (2017).
Qin, H. D. et al. Genomic characterization of esophageal squamous cell carcinoma reveals critical genes underlying tumorigenesis and poor prognosis. Am. J. Hum. Genet. 98, 709–727 (2016).
Gehring, J. S., Fischer, B., Lawrence, M. & Huber, W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics 31, 3673–3675 (2015).
Alexandrov, L. B., Nik-Zainal, S., Wedge, D. C., Campbell, P. J. & Stratton, M. R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 3, 246–259 (2013).
Forbes, S. A. et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D777–D783 (2017).
Secrier, M. et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat. Genet. 48, 1131–1141 (2017).
Weaver, J. M. J. et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat. Genet. 46, 837–843 (2014).
Petljak, M. et al. Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell 176, 1282–1294 (2019).
Seplyarskiy, V. B., Andrianova, M. A. & Bazykin, G. A. APOBEC3A/B-induced mutagenesis is responsible for 20% of heritable mutations in the TpCpW context. Genome Res. 27, 175–184 (2017).
Law, E. K. et al. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci. Adv. 2, e1601737 (2016).
Sieuwerts, A. M. et al. Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm. Cancer 5, 405–413 (2014).
Chan, T. A. et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann. Oncol. 30, 44–56 (2019).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Mularoni, L., Sabarinathan, R., Deu-Pons, J., Gonzalez-Perez, A. & Lopez-Bigas, N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 17, 128 (2016).
Rojo de la Vega, M., Chapman, E. & Zhang, D. D. NRF2 and the hallmarks of cancer. Cancer Cell 34, 21–43 (2018).
Shibata, T. et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA 105, 13568–13573 (2008).
Shibata, T. et al. NRF2 mutation confers malignant potential and resistance to chemoradiation therapy in advanced esophageal squamous cancer. Neoplasia 13, 864–873 (2011).
Totoki, Y. et al. Unique mutation portraits and frequent COL2A1 gene alteration in chondrosarcoma. Genome Res. 24, 1411–1420 (2014).
Yu, J. et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut 64, 636–645 (2015).
Papatriantafyllou, M. Cytokines: IRF7 lost in translation. Nat. Rev. Immunol. 13, 221 (2013).
Mishima, Y. et al. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 118, 2443–2453 (2011).
Weinhold, N., Jacobsen, A., Schultz, N., Sander, C. & Lee, W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 46, 1160–1165 (2014).
Adriaens, C. et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 22, 861–868 (2016).
Kim, J. et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50, 1705–1715 (2018).
Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. https://doi.org/10.1200/PO.17.00011 (2017).
Sanchez-Vega, F. et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 173, 321–337 (2018).
Deng, N. et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut 61, 673–684 (2012).
Sawada, G. et al. Genomic landscape of esophageal squamous cell carcinoma in a Japanese population. Gastroenterology 150, 1171–1182 (2016).
Gupta, R., Hong, D., Iborra, F., Sarno, S. & Enver, T. NOV (CCN3) functions as a regulator of human hematopoietic stem or progenitor cells. Science 316, 590–593 (2007).
Nusse, R. & Clevers, H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017).
Nicolai Meinshausen, P. B. H. Stability selection. J. R. Statist. Soc. B 72, 417–473 (2010).
Green, A. M. et al. Cytosine deaminase APOBEC3A sensitizes leukemia cells to inhibition of the DNA replication checkpoint. Cancer Res. 77, 4579–4588 (2017).
Nikkila, J. et al. Elevated APOBEC3B expression drives a kataegic-like mutation signature and replication stress-related therapeutic vulnerabilities in p53-defective cells. Br. J. Cancer 117, 113–123 (2017).
Mina, M. et al. Conditional selection of genomic alterations dictates cancer evolution and oncogenic dependencies. Cancer Cell 32, 155–168 (2017).
Unni, A. M., Lockwood, W. W., Zejnullahu, K., Lee-Lin, S. Q. & Varmus, H. Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. Elife 4, e06907 (2015).
Blakely, C. M. et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat. Genet. 49, 1693–1704 (2017).
Stuhlmiller, T. J. et al. Inhibition of lapatinib-induced kinome reprogramming in ERBB2-positive breast cancer by targeting BET family bromodomains. Cell Rep. 11, 390–404 (2015).
Frankell, A. M. et al. The landscape of selection in 551 esophageal adenocarcinomas defines genomic biomarkers for the clinic. Nat. Genet. 51, 506–516 (2019).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows−Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Weber, J. A., Aldana, R., Gallagher, B. D. & Edwards, J. S. Sentieon D. N. A. pipeline for variant detection-Software-only solution, over 20× faster than GATK 3.3 with identical results. PeerJ PrePrints https://doi.org/10.7287/peerj.preprints.1672v2 (2016).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Roberts, S. A. et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45, 970–976 (2013).
Mayakonda, A., Lin, D. C., Assenov, Y., Plass, C. & Koeffler, H. P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 28, 1747–1756 (2018).
Xi, R., Lee, S., Xia, Y., Kim, T. M. & Park, P. J. Copy number analysis of whole-genome data using BIC-seq2 and its application to detection of cancer susceptibility variants. Nucleic Acids Res. 44, 6274–6286 (2016).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Cheng, C. et al. Whole-genome sequencing reveals diverse models of structural variations in esophageal squamous cell carcinoma. Am. J. Hum. Genet. 98, 256–274 (2016).
Shimada, Y., Imamura, M., Wagata, T., Yamaguchi, N. & Tobe, T. Characterization of 21 newly established esophageal cancer cell lines. Cancer 69, 277–284 (1992).
Acknowledgements
A special thanks to Mr. Yanhong Li (Baidu) for his generous support of this project. We would like to thank Drs. Y. Shimada (Kyoto University), Enmin Li (Medical College of Shantou University) and Dan Su (Zhejiang Cancer Hospital) for kindly providing cell lines. This study was also supported by funding from the National Key R&D Program of China (2016YFC1302100), the CAMS Innovation Fund for Medical Sciences (2016-I2M-1-001, 2019-I2M-1-003), the National Natural Science Foundation of China (81490753, 81330063, 11971039), the fund of “San-ming” Project of Medicine in Shenzhen (No. SZSM201812088), and the Guangdong Basic and Applied Basic Research Foundation (2019B030302012).
Author information
Authors and Affiliations
Contributions
Z.L., Yanhong L. and Q.Z. supervised the study, designed the experiments, and edited the manuscript. Y.C. supervised all data analyses and experimental studies, and wrote the manuscript. H. Chen, Y. Zhao and F.H. conceived the experimental studies and performed laboratory analyses. H. Chen interpreted the functional results and reviewed the manuscript. Y. Zhao, F.H., Yang L., X.Z. and X.Y. performed the functional experiments. R.X. oversaw the bioinformatics and statistics analyses, interpreted the results and wrote and reviewed the manuscript. H. Cui, Jiawei W., Y. Zhou, W.Z., Jie W. and B.W. provided medical records and survival data, performed bioinformatics analyses, validation of variations, and statistics analyses. T.Y., E.X., X.L., Y.M., S.G., L.Z., Y.X., R.S., B.Y. and X.C. provided clinical samples and data. T.Y., P.K., Y.M., J.L. and B.S. reviewed the histopathology. T.Y., E.X., P.K., Y.M., Y. Liu., F.W., J.Y., H.L., Y.W. and Y. Zhang prepared microneedle-punctured FFPE tissues. Y. Zhai and C.C. performed the immunohistochemistry analyses.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Cui, Y., Chen, H., Xi, R. et al. Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma. Cell Res 30, 902–913 (2020). https://doi.org/10.1038/s41422-020-0333-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-020-0333-6
This article is cited by
-
The genomic landscape of esophageal squamous cell carcinoma cell lines
Cancer Cell International (2025)
-
Multi-omics analysis reveals immunosuppression in oesophageal squamous cell carcinoma induced by creatine accumulation and HK3 deficiency
Genome Medicine (2025)
-
Integrative analysis of T cell-mediated tumor killing-related genes reveals KIF11 as a novel therapeutic target in esophageal squamous cell carcinoma
Journal of Translational Medicine (2025)
-
Survival outcome prediction of esophageal squamous cell carcinoma patients based on radiomics and mutation signature
Cancer Imaging (2025)
-
A Risk Prognostic Signature Basing on Cuproptosis-related Ferroptosis and Immune Genes for Esophageal Squamous Cell Carcinoma Patients
Cell Biochemistry and Biophysics (2025)