Abstract
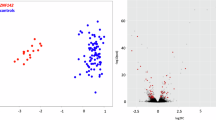
Developmental Delay with Gastrointestinal, Cardiovascular, Genitourinary, and Skeletal Abnormalities syndrome (DEGCAGS, MIM #619488) is caused by biallelic, loss-of-function (LoF) ZNF699 variants, and is characterized by variable neurodevelopmental disability, discordant organ anomalies among full siblings and infant mortality. ZNF699 encodes a KRAB zinc finger protein of unknown function. We aimed to investigate the genotype-phenotype spectrum of DEGCAGS and the possibility of a diagnostic DNA methylation episignature, to facilitate the diagnosis of a highly variable condition lacking pathognomonic clinical findings. We collected data on 30 affected individuals (12 new). GestaltMatcher analyzed fifty-three facial photographs from five individuals. In nine individuals, methylation profiling of blood-DNA was performed, and a classification model was constructed to differentiate DEGCAGS from controls. We expand the ZNF699-related molecular spectrum and show that biallelic, LoF, ZNF699 variants cause unique clinical findings with age-related presentation and a similar facial gestalt. We also identified a robust episignature for DEGCAGS syndrome. DEGCAGS syndrome is a clinically variable recessive syndrome even among siblings with a distinct methylation episignature which can be used as a screening, diagnostic and classification tool for ZNF699 variants. Analysis of differentially methylated regions suggested an effect on genes potentially implicated in the syndrome’s pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data and materials can be supplied upon request. All seven novel ZNF699 variants were deposited in ClinVar, accession numbers SCV004565351 – SCV004565357. Some of the datasets used in this study are publicly available and may be obtained from gene expression omnibus (GEO) using the following accession numbers. GEO: GSE116992, GSE66552, GSE74432, GSE97362, GSE116300, GSE95040, GSE104451, GSE125367, GSE55491, GSE108423, GSE116300, GSE 89353, GSE52588, GSE42861, GSE85210, GSE87571, GSE87648, GSE99863, and GSE35069. These include DNA methylation data from individuals with Kabuki syndrome, Sotos syndrome, CHARGE syndrome, immunodeficiency-centromeric instability-facial anomalies (ICF) syndrome, Williams-Beuren syndrome, Chr7q11.23 duplication syndrome, BAFopathies, Down syndrome, a large cohort of unresolved subjects with developmental delays and congenital abnormalities, and also several large cohorts of DNA methylation data from the general population. Remaining data are not available due to institutional or REB restrictions. EpiSign is proprietary, trademarked analytical software owned by EpiSign Inc. and parts of it are based on the methods and publicly available software that are referenced in the methods section.
References
Bertoli-Avella AM, Kandaswamy KK, Khan S, Ordonez-Herrera N, Tripolszki K, Beetz C, et al. Combining exome/genome sequencing with data repository analysis reveals novel gene-disease associations for a wide range of genetic disorders. Genet Med. 2021;23:1551–68.
Biela M, Rydzanicz M, Jankowska A, Szlagatys-Sidorkiewicz A, Rozensztrauch A, Ploski R, et al. Further delineation of developmental delay with gastrointestinal, cardiovascular, genitourinary, and skeletal abnormalities caused by ZNF699 gene mutation. Genes (Basel). 2022;13:168.
de Tribolet-Hardy J, Thorball CW, Forey R, Planet E, Duc J, Coudray A, et al. Genetic features and genomic targets of human KRAB-zinc finger proteins. Genome Res. 2023;33:1409–23.
Rosspopoff O, Trono D. Take a walk on the KRAB side: (Trends in Genetics, 39:11 p:844-57, 2023). Trends Genet. 2023.
Levy MA, McConkey H, Kerkhof J, Barat-Houari M, Bargiacchi S, Biamino E, et al. Novel diagnostic DNA methylation episignatures expand and refine the epigenetic landscapes of Mendelian disorders. HGG Adv. 2022;3:100075.
Sadikovic B, Levy MA, Kerkhof J, Aref-Eshghi E, Schenkel L, Stuart A, et al. Clinical epigenomics: genome-wide DNA methylation analysis for the diagnosis of Mendelian disorders. Genet Med. 2021;23:1065–74.
Aref-Eshghi E, Kerkhof J, Pedro VP, Groupe DIF, Barat-Houari M, Ruiz-Pallares N, et al. Evaluation of DNA methylation episignatures for diagnosis and phenotype correlations in 42 Mendelian neurodevelopmental disorders. Am J Hum Genet. 2020;106:356–70.
Kerkhof J, Rastin C, Levy MA, Relator R, McConkey H, Demain L, et al. Diagnostic utility and reporting recommendations for clinical DNA methylation episignature testing in genetically undiagnosed rare diseases. Genet Med. 2024;26:101075.
Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–30.
Pranav Chand R, Vinit W, Vaidya V, Iyer AS, Shelke M, Aggarwal S, et al. Proband only exome sequencing in 403 Indian children with neurodevelopmental disorders: Diagnostic yield, utility and challenges in a resource-limited setting. Eur J Med Genet. 2023;66:104730.
Saleh S, Beyyumi E, Al Kaabi A, Hertecant J, Barakat D, Al Dhaheri NS, et al. Spectrum of neuro-genetic disorders in the United Arab Emirates national population. Clin Genet. 2021;100:573–600.
Hsieh TC, Bar-Haim A, Moosa S, Ehmke N, Gripp KW, Pantel JT, et al. GestaltMatcher facilitates rare disease matching using facial phenotype descriptors. Nat Genet. 2022;54:349–57.
Hustinx AHF, Sümer O, Javanmardi B, André E, Peter Krawitz P, Hsieh Tz-Ch. Improving deep facial phenotyping for ultra-rare disorder verification using model ensembles. IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA. 2023:5007-17.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, et al. ClinGen—the clinical genome resource. N Engl J Med. 2015;372:2235–42.
Abou Tayoun AN, Pesaran T, DiStefano MT, Oza A, Rehm HL, Biesecker LG, et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat 2018;39:1517–24.
Jarvik GP, Browning BL. Consideration of cosegregation in the pathogenicity classification of genomic variants. Am J Hum Genet. 2016;98:1077–81.
Aref-Eshghi E, Rodenhiser DI, Schenkel LC, Lin H, Skinner C, Ainsworth P, et al. Genomic DNA methylation signatures enable concurrent diagnosis and clinical genetic variant classification in neurodevelopmental syndromes. Am J Hum Genet. 2018;102:156–74.
Aref-Eshghi E, Kerkhof J, Pedro VP, France GD, Barat-Houari M, Ruiz-Pallares N, et al. Evaluation of DNA methylation episignatures for diagnosis and phenotype correlations in 42 Mendelian neurodevelopmental disorders. Am J Hum Genet. 2021;108:1161–3.
Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–9.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinforma. 2012;13:86.
Benjamini YAYH. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B (Methodol). 1995;57:289–300.
Levy MA, Relator R, McConkey H, Pranckeviciene E, Kerkhof J, Barat-Houari M, et al. Functional correlation of genome-wide DNA methylation profiles in genetic neurodevelopmental disorders. Hum Mutat 2022;43:1609–28.
Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, Lord RV, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6.
Cavalcante RG, Sartor MA. annotatr: genomic regions in context. Bioinformatics. 2017;33:2381–3.
Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics. 2016;32:286–8.
Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–67.
Alghanim H, Antunes J, Silva D, Alho CS, Balamurugan K, McCord B. Detection and evaluation of DNA methylation markers found at SCGN and KLF14 loci to estimate human age. Forensic Sci Int Genet. 2017;31:81–8.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9.
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. Alphafold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D44.
Ali SM, AlMasri DA, Prada CE, Lin D, Bosley TM, Kozak I. Clinical and ocular abnormalities in DEGCAGS syndrome-Developmental delay with gastrointestinal, cardiovascular, genitourinary, and skeletal abnormalities. Mol Genet Genom Med. 2024;12:e2329.
Abraham KJ, Khosraviani N, Chan JNY, Gorthi A, Samman A, Zhao DY, et al. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature. 2020;585:298–302.
Al-Naama N, Mackeh R, Kino T. C(2)H(2)-type zinc finger proteins in brain development, neurodevelopmental, and other neuropsychiatric disorders: systematic literature-based analysis. Front Neurol. 2020;11:32.
Lesmann H, Klinkhammer H, Pdmdpp MK. The future role of facial image analysis in ACMG classification guidelines. Med Genet. 2023;35:115–21.
Hennekam RCM. Pathophysiology of premature aging characteristics in Mendelian progeroid disorders. Eur J Med Genet. 2020;63:104028.
Hennekam RCM. The external phenotype of aging. Eur J Med Genet. 2020;63:103995.
Bai X, Bian Z. MicroRNA-21 is a versatile regulator and potential treatment target in central nervous system disorders. Front Mol Neurosci. 2022;15:842288.
Peeney D, Liu Y, Lazaroff C, Gurung S, Stetler-Stevenson WG. Unravelling the distinct biological functions and potential therapeutic applications of TIMP2 in cancer. Carcinogenesis. 2022;43:405–18.
He X, Chen Z, Jiang Y, Qiu X, Zhao X. Different mutations of the human c-mpl gene indicate distinct haematopoietic diseases. J Hematol Oncol. 2013;6:11.
Riley BP, Kalsi G, Kuo PH, Vladimirov V, Thiselton DL, Vittum J, et al. Alcohol dependence is associated with the ZNF699 gene, a human locus related to Drosophila hangover, in the Irish Affected Sib Pair Study of Alcohol Dependence (IASPSAD) sample. Mol Psychiatry. 2006;11:1025–31.
Ali MA, Way MJ, Marks M, Guerrini I, Thomson AD, Strang J, et al. Phenotypic heterogeneity in study populations may significantly confound the results of genetic association studies on alcohol dependence. Psychiatr Genet. 2015;25:234–40.
Cacace R, Heeman B, Van Mossevelde S, De Roeck A, Hoogmartens J, De Rijk P, et al. Loss of DPP6 in neurodegenerative dementia: a genetic player in the dysfunction of neuronal excitability. Acta Neuropathol. 2019;137:901–18.
Maussion G, Cruceanu C, Rosenfeld JA, Bell SC, Jollant F, Szatkiewicz J, et al. Implication of LRRC4C and DPP6 in neurodevelopmental disorders. Am J Med Genet A 2017;173:395–406.
Lewis EM, Stein-O’Brien GL, Patino AV, Nardou R, Grossman CD, Brown M, et al. Parallel social information processing circuits are differentially impacted in autism. Neuron. 2020;108:659–75.e6.
Anvar LH, Alejafar A, Moosavi SE, Charsouei S, Zeynalzadeh N, Fanid LM, et al. The study of rs324420 (C385A) polymorphism of the FAAH gene of the endocannabinoid system in patients with epilepsy and ADHD. Epilepsy Res. 2023;192:107100.
Flaherty E, Maniatis T. The role of clustered protocadherins in neurodevelopment and neuropsychiatric diseases. Curr Opin Genet Dev. 2020;65:144–50.
Laufer BI, Chater-Diehl EJ, Kapalanga J, Singh SM. Long-term alterations to DNA methylation as a biomarker of prenatal alcohol exposure: From mouse models to human children with fetal alcohol spectrum disorders. Alcohol. 2017;60:67–75.
Acknowledgements
We are grateful to the participating families. This work has been generated within the Norwegian National Advisory Unit on Rare Disorders Project 2629394, ‘The post-exome clinic: improving the impact of exome sequencing for developmental disorders in Norway’ and the European Reference Network on Rare Congenital Malformations and Rare Intellectual Disability (ERN-ITHACA) [EU Framework Partnership Agreement ID: 3HP-HP-FPA ERN-01-2016/739516]. The GestaltMatcher database is a service operated by the Association for Genome Diagnostics (AGD), which is a registered non-for-profit organization in Germany. GMDB aims to improve the openness and accessibility of scientific findings and to enhance collaboration amongst researchers and clinicians. GMDB is a non-profit community resource and is not linked to any one publisher or journal. We thank graphic designer Rannveig Lohne for assisting in creating Fig. 1a.
Funding
This study was funded by the Norwegian National Advisory Unit on Rare Disorders (grant #43066 to SDH), the government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-188 to BS), grants from the National Institutes of Health (DK068306) and the Begg Family Foundation (both to FH) and grants by the Interdisciplinary Center for Clinical Research Erlangen (J70) and the Deutsche Forschungsgemeinschaft (281319475) (both to TJS). Sequencing and analysis of [GAZ_431/ Patient 16] were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and were funded by the National Human Genome Research Institute (NHGRI) grants UM1HG008900 (with additional support from the National Eye Institute, and the National Heart, Lung and Blood Institute), and R01HG009141 (to LP). All funders played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: DW, BS and SDH; Data Curation: KK, MH, IA, T-ChHs, HL., AA and DW; Formal Analysis: KK, DW, IA, and T-ChHs; Funding acquisition: FH, TJS, BS and SDH; Investigation: KK, DW, IA, T-ChHs, JP, RML, LD, MD, HL, TL, KHS, B.M.S.Al-M, RGYAl-Ob, DM, MR, MB, MSS, AA, HTG, LP, ShSh, HD, SB, FL, AS, TK, TJ-Sch, NAS, RR, MAL, JK, JS, HH, GVP, FH, SBS, RM, TWY, and SDH; Methodology: KK, T-ChHs, MAL, BS and SDH; Project Administration: HL, RM, MH, DW, KK, IA and SDH; Software: KK, IA, T-ChHs, BS and PK; Supervision: DW, RML, SBS, RM, TY, PK, BS and SDH; Validation: KK, IA, T-ChHs; Visualization: KK, DW, IA, RML, LD, T-ChHs and SDH; Writing-original draft: KK, DW, T-ChHs, IA and SDH; Writing-review and editing: KK, DW, T-ChHs, AA, BS and SDH.
Corresponding authors
Ethics declarations
Competing interests
BS is a shareholder in EpiSign Inc, company involved in commercialization of EpiSignTM technology.
Ethical approval
This study was performed according to the Declaration of Helsinki and approved by the Western Norway Regional Ethics Committee (REC 604007) and the Western Ontario University Research Ethics Board (REB 106302). Written informed consent for the publication of photographs, videos and medical information was obtained from parents/legal guardians.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Karimi, K., Weis, D., Aukrust, I. et al. Epigenomic and phenotypic characterization of DEGCAGS syndrome. Eur J Hum Genet 32, 1574–1582 (2024). https://doi.org/10.1038/s41431-024-01702-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-024-01702-y
This article is cited by
-
Biallelic loss-of-function variants in ZNF142 are associated with a robust DNA methylation signature affecting a limited number of genomic loci
European Journal of Human Genetics (2025)
-
New guidelines for rare cancer syndromes
European Journal of Human Genetics (2024)