Abstract
Breast cancer remains a significant global health challenge. In Australia, the adoption of publicly-funded multigene panel testing for eligible cancer patients has increased accessibility to personalised care, yet has also highlighted the increasing prevalence of variants of uncertain significance (VUS), complicating clinical decision-making. This project aimed to explore the spectrum and actionability of breast cancer VUS in Australian familial cancer centers (FCCs). Leveraging data from 11 FCCs participating in the Inherited Cancer Connect database, we retrieved VUS results from 1472 patients. Through ClinVar crosschecks and application of gene-specific ACMG/AMP guidelines, we showed the potential for reclassification of 4% of unique VUS as pathogenic or likely pathogenic, and 80% as benign or likely benign. Surveys conducted with FCCs and diagnostic laboratories described current practices and challenges in variant reclassifications, highlighting resource constraints preventing periodic VUS review and notifications from the laboratories to the FCCs. Our study suggests there are benefits to routine VUS review and reclassification, particularly in publicly-funded healthcare systems. Future research should focus on assessing the clinical impact and cost-effectiveness of implementing routine variant review practices, alongside efforts to enhance communication between FCCs and laboratories.
Similar content being viewed by others
Introduction
Breast cancer is the most prevalent cancer diagnosed in women worldwide. The proportion of breast cancers in the general population attributed to BRCA1 and BRCA2 germline pathogenic variants is approximately 5–10% [1, 2], but this proportion is much higher (up to 55%) in selected families referred to familial cancer clinics [3]. Even though rarer than BRCA1 and BRCA2, a short list of other highly penetrant genes are known to elevate breast cancer risk [4]. In general, hereditary breast cancer is associated with poorer survival rates compared to breast cancer as a whole [5, 6].
Genetic testing is valuable as it provides patients and their close relatives with knowledge about their hereditary cancer risk, and guides personalised strategies for cancer prevention and treatment for both pathogenic variant carriers and those with “normal” results (no germline pathogenic variant identified). Multigene panel testing has been adopted as a cost-effective strategy to simultaneously sequence multiple genes; however, these panels reveal variants of uncertain significance (VUS) more frequently than single gene testing approaches. Identification of VUS presents an important challenge to the clinical utility of precision genomic medicine [7]. These variants not only limit effective clinical management but are also a source of anxiety and confusion for some clinicians, and for patients and their family members [8]. Presently, there are no definitive recommendations regarding the reporting of VUS, and attitudes on their disclosure vary among clinical and laboratory genetic counsellors [9].
Testing of breast cancer predisposition genes increased significantly in Australia following the introduction of national government funding of multigene panel testing in 2017. The uptake of multigene panel testing is expected to have led to a corresponding increase in the number of breast cancer gene VUS identified [10]. ClinVar is a comprehensive resource that aggregates variant data from multiple sources, including clinical laboratories, and thus provides a useful overview of the diversity and classification of variants encountered in clinical and research settings [11]. For the subset of genes whose testing is publicly funded in Australia (BRCA1, BRCA2, CDH1, PALB2, PTEN, TP53), the proportion of unique variants in ClinVar with aggregate clinical interpretation as VUS ranges from 16% to (BRCA1) to 47% (CDH1) (ClinVar, accessed 21 April 2024). Furthermore, when variants with conflicting classifications are taken into account, these two categories collectively constitute at least a third of all the variants submitted to ClinVar for each gene.
VUS reclassification has potential to alter clinical management in hereditary cancer patients [12]; however, little is known about the prevalence and potential for VUS reclassifications among breast cancer patients. The rate of VUS reclassification from purposeful variant reclassification efforts using American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) guidelines was 61% for a study of 92 patients with any VUS received through clinical testing [13], with ranges of up to 81.3% for BRCA1/2-focused studies [14,15,16,17,18]. It is therefore reasonable to think that reclassification of VUS in breast cancer risk genes has potential to inform patient clinical management.
The aim of this project was to identify the scope of the problem of breast cancer VUS in Australian familial cancer centres (FCCs), and to understand reclassification processes and interaction between FCCs and diagnostic laboratories, in order to consider future strategies for increased actionability and improved clinical management of breast cancer patients.
Materials and methods
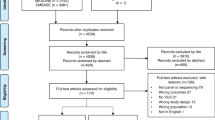
An overview of the study process is shown in Fig. 1.
Data acquisition
We engaged with 11 Australian FCCs participating in the Inherited Cancer Connect (ICCon) partnership (Forrest, 2018) to retrieve the breast cancer gene variants listed as VUS in their internal databases in the following genes eligible for publicly-funded testing in Australia: BRCA1, BRCA2, CDH1, PALB2, PTEN, and TP53. The participating FCCs were: ACT Genetic Service, Austin Health, the Adult Genetics Unit (a state-wide service for South Australia), Genetic Health Queensland, Genetic Health WA, Liverpool Cancer Genetics, Peter MacCallum Cancer Centre, Princes of Wales Hospital, Royal Melbourne Hospital, Royal North Shore, and St. George Hospital. Data retrieved corresponded to VUS results for patients undergoing multigene panel testing due to hereditary cancer risk identified post-2017 and up to 2022.
For the six selected genes, we received VUS results from 1516 patients. Patient data records, stored in the clinical databases of the FCCs using proprietary software (such as Progeny, TrakGene, FamBIS) were internally queried by FCC data managers using a database search function for terms to identify variants for any patient annotated with classification interpreted to be VUS in one of the genes of interest; the specific terms were selected according to the data dictionary for the relevant FCC, e.g., Class 3, VUS, VOUS. We first checked for nomenclature errors using the Mutalyzer tool [19]. Entries for 44 patients were unresolvable and were excluded, leaving a total of 1472 patients available for analyses. The nomenclature for the remaining entries was standardized as per HGVS recommendations.
ClinVar crosscheck and groupings
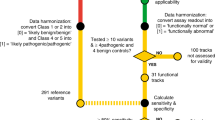
To ascertain the extent to which variants labelled as “VUS” in FCC internal databases may have resulted from outdated or inadequately reassessed historical reports, we initially examined their classifications in ClinVar (as at September 2022). These variants were categorised into three distinct groups, as follows:
-
Variants with Expert Panel submissions as Likely pathogenic and Pathogenic (hereafter referred to as P/LP) or Likely benign and Benign (hereafter referred to as B/LB)
-
Variants with multiple non-conflicting submissions as P/LP or B/LB
-
Variants not belonging to any of these groups (i.e. variants with a single submission as P/LP or B/LB, variants with conflicting submissions, variants classified as Uncertain, and variants not previously submitted to ClinVar), hereafter referred to as “Other”
Gene-specific ACMG/AMP guidelines application
We applied gene-specific ACMG/AMP criteria to variants in the “Other” ClinVar group, using the following criteria specifications versions: BRCA1 v1.1.0, BRCA2 v1.1.0, CDH1 v3.1.0, PALB2 v1.1.0, PTEN v3.0.0, and TP53 v2.0.0. Details for specifications are available in the ClinGen Criteria Specification Registry (https://cspec.genome.network/cspec/ui/svi/). For the population-based codes (BA1, BS1, PM2), we used the gnomAD v4.1 database [20], except for BRCA1 and BRCA2 for which we performed this step prior to v4 release, and whose latest guidelines specify the use of gnomAD v2.1 and gnomAD v3.1 (non-cancer datasets). For all genes, the cutoffs used for SpliceAI splicing predictions were ≥0.2 (predicted impact) and ≤0.1 (no predicted impact), as recommended by the ClinGen Sequence Variant Interpretation Splicing Subgroup [21]. All other codes were assigned following the respective gene specifications. The proposed class following review was assigned based on a Bayesian point systems [22], which included reclassifying as LB variants with a total of −1 points where the only pathogenic code applied is PM2_Supporting, and there are at least two benign codes applied.
Surveys to FCCs and diagnostic laboratories
To enhance comprehension of the VUS review process and the dynamics between FCCs and diagnostic laboratories in Australia, as well as to identify unmet clinical needs, we developed two surveys:
i) An eight-question survey was sent to each FCC representative, totalling nine representatives from 11 FCCs. The survey aimed to gain insights into how FCCs become aware of VUS reclassifications, actions taken post-notification of VUS upgrades/downgrades, and reasons for failing to update their internal clinical database.
ii) A 13-question survey was sent to each laboratory designated as being used by the FCCs, totalling six representatives from six laboratories. This survey sought to understand the reasons prompting VUS review, the frequency and constraints of VUS review, how laboratories learn of new Expert Panel classifications and subsequent actions, notifications of reclassifications to FCCs, and perspectives on the significance of regular VUS review.
Both of these surveys (Supplementary Material 1) comprised multiple-choice questions, all of which had an option to provide additional comments when considered relevant, as well as separate open-ended questions. Multiple choice responses were analysed by frequency, and the most frequent responses to open-ended responses were summarised.
Results
Variant spectrum per gene and ClinVar group
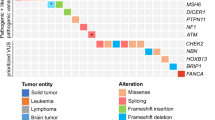
Data was available from 1472 patient records for breast cancer gene variants from all 11 participating FCCs, corresponding to 944 unique variants recorded as “VUS” across the FCC internal databases. Of these, the number of VUS per gene was as follows: 522 in BRCA2, 265 in BRCA1, 104 in PALB2, 37 in TP53, 11 in PTEN, and five in CDH1.
An overview of the results from the ClinVar crosscheck is shown in Fig. 2. At the time (September 2022), approximately 20% of the unique VUS had Expert Panel submissions as P/LP [11] and B/LB (175), while 5% had multiple non-conflicting submissions in the same directions (9P/LP and 36 B/LB). For each gene, most of the VUS were in the “Other” group.
Results from the ClinVar crosscheck were returned to FCC representatives prior to the surveys, for consideration of possible reclassification review; in particular variants in ClinVar with Expert Panel assertion other than VUS, or with multiple non-conflicting submissions.
Gene-specific ACMG/AMP application
We applied gene-specific ACMG/AMP guidelines to all the VUS in the “Other” ClinVar group (total 713 variants). The number of our resulting proposed reclassifications per gene is shown in Fig. 3. There was no suggested reclassification for three CDH1 variants. Application of the specifications for the remaining genes overall reclassified 3% of all variants as P/LP (20 out of the 713 “Other” VUS), and 77% variants as B/LB (546). These variants, codes applied, and our suggested classification are detailed in Supplementary Table 1. As an update, we added updated ClinVar classification details (as at 27 January 2024).
Results from FCC survey
We received survey results from representatives of all 11 participating FCCs, with some clinicians answering on behalf of multiple centres. A simplified summary of key survey results is shown in Table 1.
Regarding how FCCs become aware of VUS reclassifications, four of the nine FCC representatives reported that their laboratory provider routinely notifies them of VUS upgrades, while this number decreased to three for downgrades. Other methods selected were receiving a diagnostic report for a new patient with the same variant (3/9) and conducting regular reviews of variants within their internal clinical database (2/9). Additionally, some FCC representatives provided free-text responses citing additional reasons such as patients reaching out for more information (3), referral of a relative (3), and involvement in local and/or national multidisciplinary team (MDT) meetings (1).
As per actions taken upon VUS reclassifications, it was found that all participating FCCs requested the re-issue of reports for patients in cases of VUS upgrades, whereas only six out of nine FCC representatives did so for variant downgrades.
Lastly, the majority of FCC representatives reported that the primary reason for the lack of updates in their internal clinical databases was their unawareness of VUS reclassifications (8/9), with a lack of resources also cited as a contributing factor by a third of the representatives (3/9), and not considered clinically necessary by one representative (1/9).
Results from laboratory survey
We received survey results from representatives of all six participating laboratories. A summary of the responses regarding activities prompting review of previously identified VUS is presented in Table 2, with a scale provided from 1 (least often) to 5 (most often) for the activities considered relevant by the laboratories.
The activity most commonly cited as prompting review was FCCs contacting laboratories for additional information (average rating of 3.83), followed by laboratories identifying new evidence (average rating of 3). Conversely, activities least likely to prompt VUS review included laboratories generating or accessing external research findings (average rating of 0.7), followed by the availability of new ClinVar submissions (average rating of 0.8).
A simplified summary of other key survey results from the six laboratory representatives is shown in Table 3. Only two out of six laboratories reported conducting regular reviews of VUS, one annually, the other at least biannually. Lack of resources was cited by all laboratories as a primary reason for not regularly conducting this activity. Additional reasons noted by half of the laboratories were an inability to track previous results for variants not included in reports, and the activity not deemed clinically relevant unless prompted by FCCs for additional information.
Regarding awareness of new Expert Panel reclassifications in an open-ended survey question, two out of the six laboratories indicated that they had easier access to these updates by virtue of being Variant Curation Expert Panel (VCEP) members, while the remaining laboratories lacked a standardised process.
Another open-ended survey question revealed the following actions taken following new Expert Panel reclassifications, including: review and/or discussion of provided information for potential reclassification (3); reclassification of relevant variants (1); and informing the relevant FCC to request a reclassification from the lab (1).
With regards to notifications to FCCs, all laboratories reported notifying FCCs of VUS upgrades, compared to five out of six for downgrades. One of the laboratories reporting notifications of both options added a note to clarify that these were expected actions, since they would only review VUS following a previous FCC enquiry. The majority of laboratories reported having a documented process in place for notifying FCCs of VUS reclassifications (5/6).
Lastly, all laboratories except one indicated that VUS regular review would be prioritised if additional funding were available (5/6).
Discussion
We have performed a nation-wide audit of VUS recorded in the clinical databases of Australian familial cancer services to determine the scale of unrecognised actionability, and to map the current and required processes to achieve effective variant review. Our ultimate aim is to inform future improvements in Australian practices concerning variant classification, reducing variant uncertainty and improving patient clinical management.
With a straightforward crosscheck of the 944 clinically detected VUS retrieved from FCCs against ClinVar, we found that a notable number of the historical reports recorded in FCC internal clinical databases had not undergone updates and/or reassessment with existing new evidence. Approximately a quarter of the so-called VUS had an Expert Panel or multiple non-conflicting submissions as P/LP or B/LB, information that was returned to the FCCs to encourage further review of these variants. For the larger group of remaining variants that are uncertain, have single or conflicting submissions in ClinVar or that are absent from the database altogether (n = 713), we found that application of gene-specific VCEP guidelines resulted in a 79% reduction in the VUS rate, even when relying solely on publicly available information. When combining both results, our analysis suggests that 84% of the initially provided 944 unique VUS could potentially be reclassified as P/LP or B/LB. While most of these potential reclassifications were in the benign direction (80%), there are many reasons to consider variant downgrades as clinically important, including the opportunity to avoid unnecessary clinical risk management interventions or relief from any continuing anxiety, although it can also be an indication to continue searching for alternative genetic explanations.
To better understand the factors contributing to these findings, and to describe the barriers to FCCs and laboratory identifying and implementing reclassifications, we surveyed both groups. Representatives from a large majority of FCCs (89%) indicated that they do not routinely update their internal variant database as they are generally not aware of any potential reclassifications. Additional FCC survey results indicated that 100% of VUS upgrades and 67% of downgrades, would be communicated to patients. In general, this attitude aligns with findings from a US-based survey study where both oncologists and genetic counsellors concurred that all reclassified variants, even when they do not represent a change in clinical management, should be disclosed to patients [23]. On the other hand, in the laboratory survey results, laboratories indicated that they notify the FCCs of variant reclassifications in nearly all instances, acknowledging that the primary reason prompting VUS review was an FCC request to the laboratory for additional or updated information. This highlights a major flaw in the current VUS review process: FCCs heavily rely on laboratories to keep them updated of variant reclassifications, as they do not have the clinical capacity and/or expertise to do their own reviews, but laboratories rarely initiate VUS review without a clinical prompt. The results suggest that only a relatively low proportion of VUS found in patients will be put forward for further review due to a request from the FCC or as a result of a relative undergoing testing, and that in most instances where a potential reclassification could occur based on updated information and/or classification methods, this opportunity is missed. Only two of the six surveyed laboratories indicated that they initiate their own variant re-review on a regular basis. Resources was the unanimous reason for no more regular or initiated variant re-review, and all but one of the laboratories noted that they would prioritise VUS review if they had more funding to do so. One laboratory representative elaborated that regular VUS review, regardless of the funding, requires significant consideration of the downstream clinical interactions and interventions, and highlighted concerns about possible legal implications from unrecognised actionability of VUS.
These results are a prompt to consider how a more effective and reliable process could be implemented for identifying reclassifiable VUS in clinical datasets and communicating updated curations to facilitate optimal care for patients. A simple and affordable recommendation to streamline the VUS review process between clinical services and laboratories is to promote automated notifications to the laboratories of Expert Panel reclassifications for variants within their curation system. In Australia, there is potential to facilitate this by leveraging the properties of Shariant [24], a system for real-time sharing of variant data between Australian FCCs and laboratories, which has recently introduced this capability. This could be supplemented by general look-ups to ClinVar, preferably in an automated manner as recently proposed [25]. Ideally, this approach would be coupled with encouraging proactive notification of reclassifications by laboratories to FCCs. However, Expert Panels do not typically perform large-scale reviews of variants, as this typically exceeds the capacity of the VCEPs and is not prioritised by ClinGen, which usually focuses on variants with multiple uncertain or conflicting submissions. Therefore, while this “ClinVar notification” approach can be beneficial, it will not address the larger number of variants that are not reviewed by Expert Panels but would still benefit from more frequent laboratory-based reviews following gene-specific ACMG/AMP guidelines.
There was no clear consensus in the survey responses around which group bears the final responsibility for the task of continuing review of VUS to identify potential reclassifications. A more comprehensive solution is required for promoting periodic laboratory review of VUS for potential increased actionability. We expect that the reclassification rate of this study would be greater with inclusion of evidence held privately within laboratories. The reclassification rate is also likely to increase with evolution of ClinGen gene-specific classification guidelines, which has been demonstrated in several VCEP-related studies to reduce the VUS rate [26,27,28].
Regular VUS review and variant reclassification will require resources, adding to the routine work of trained staff. Numerous studies have demonstrated the cost-effectiveness of genetic testing in women with breast cancer [29,30,31]. In response, countries such as Australia and the UK have integrated genetic testing for hereditary breast cancer risk into their respective government funded health services (Medicare and the NHS), targeting patients meeting specific eligibility criteria, including those with a 10% risk of carrying a germline pathogenic variant based on clinical presentation. It should be noted that VUS review and variant reclassification activities to identify a P/LP variant should be less costly than the initial genetic testing that incurred the costs of the initial consultation, sample collection, laboratory analysis, sequencing bioinformatics. Costs associated with VUS review would be limited to resources for re-curation costs, and re-issue of reports when necessary. Additionally, reclassifying VUS as B/LB (80% reclassification rate as per our work) has the potential to lead to savings by avoiding the cost of unnecessary clinical interventions, recognising that not all VUS are considered clinically relevant and management might be dependent on other factors, and by removing variants from the list of VUS requiring ongoing review by the laboratory. Further, for VUS previously returned to patients, there is potential to provide reassurance and relieve anxiety following a B/LB reclassification. Therefore, publicly funding the VUS review process in countries where hereditary cancer testing is already publicly funded appears likely to have economic benefits. In terms of VUS review frequency, two of the laboratories reported conducting this activity every 1-2 years, aligning with reported most effective frequencies [32, 33]. The inconsistency in approaches between diagnostic laboratories reveals a need to develop a national approach to standardise variant reclassification protocols. The consensus framework on variant reclassification developed by CanVIG-UK [34] might provide a useful starting point in this regard.
Future directions of this work include assessing the clinical impact of our findings, and the cost-effectiveness of implementing routine variant review and reclassification practices. In parallel, national efforts should be directed towards promoting collaboration between FCCs and laboratories to ensure timely notification processes, potentially through standardised protocols and increased communication channels.
Data availability
Numerous data are made available via supplementary materials. Additional data can be made available on reasonable request.
References
Larsen MJ, Thomassen M, Gerdes AM, Kruse TA. Hereditary breast cancer: clinical, pathological and molecular characteristics. Breast Cancer. 2014;8:145–55.
Mehrgou A, Akouchekian M. The importance of BRCA1 and BRCA2 genes mutations in breast cancer development. Med J Islam Repub Iran. 2016;30:369.
Adedokun B, Zheng Y, Ndom P, Gakwaya A, Makumbi T, Zhou AY, et al. Prevalence of Inherited Mutations in Breast Cancer Predisposition Genes among Women in Uganda and Cameroon. Cancer Epidemiol Biomark Prev. 2020;29:359–67.
Daly MB, Pal T, Maxwell KN, Churpek J, Kohlmann W, AlHilli Z, et al. NCCN Guidelines® Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2024. J Natl Compr Canc Netw. 2023;21:1000–10.
Blondeaux E, Arecco L, Punie K, Graffeo R, Toss A, De Angelis C, et al. Germline TP53 pathogenic variants and breast cancer: A narrative review. Cancer Treat Rev. 2023;114:102522.
Baretta Z, Mocellin S, Goldin E, Olopade OI, Huo D. Effect of BRCA germline mutations on breast cancer prognosis: A systematic review and meta-analysis. Medicine. 2016;95:e4975.
Amano Y, Raz A, Timmermans S, Shkedi-Rafid S. Cancer patients’ understandings of genetic variants of uncertain significance in clinical care. J Community Genet. 2022;13:381–8.
Makhnoon S, Shirts BH, Bowen DJ. Patients’ perspectives of variants of uncertain significance and strategies for uncertainty management. J Genet Couns. 2019;28:313–25.
Chang EY, Solomon I, Culver JO, Gorman N, Comeaux JG, Lerman C, et al. Clinical and laboratory genetic counselor attitudes on the reporting of variants of uncertain significance for multigene cancer panels. J Genet Couns. 2023;32:706–16.
Rehm HL, Alaimo JT, Aradhya S, Bayrak-Toydemir P, Best H, Brandon R, et al. The landscape of reported VUS in multi-gene panel and genomic testing: Time for a change. Genet Med. 2023;25:100947.
Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–5.
Makhnoon S, Levin B, Ensinger M, Mattie K, Volk RJ, Zhao Z, et al. A multicenter study of clinical impact of variant of uncertain significance reclassification in breast, ovarian and colorectal cancer susceptibility genes. Cancer Med. 2023;12:2875–84.
Tsai GJ, Rañola JMO, Smith C, Garrett LT, Bergquist T, Casadei S, et al. Outcomes of 92 patient-driven family studies for reclassification of variants of uncertain significance. Genet Med. 2019;21:1435–42.
So MK, Jeong TD, Lim W, Moon BI, Paik NS, Kim SC, et al. Reinterpretation of BRCA1 and BRCA2 variants of uncertain significance in patients with hereditary breast/ovarian cancer using the ACMG/AMP 2015 guidelines. Breast Cancer. 2019;26:510–9.
Innella G, Ferrari S, Miccoli S, Luppi E, Fortuno C, Parsons MT, et al. Clinical implications of VUS reclassification in a single-centre series from application of ACMG/AMP classification rules specified for BRCA1/2. J Med Genet. 2024;61:483–9.
Ha HI, Ryu JS, Shim H, Kong SY, Lim MC. Reclassification of BRCA1 and BRCA2 variants found in ovarian epithelial, fallopian tube, and primary peritoneal cancers. J Gynecologic Oncol. 2020;31:e83.
Li D, Shi Y, Li A, Cao D, Su H, Yang H, et al. Retrospective reinterpretation and reclassification of BRCA1/2 variants from Chinese population. Breast Cancer. 2020;27:1158–67.
Liu Y, Wang H, Wang X, Liu J, Li J, Wang X, et al. Prevalence and reclassification of BRCA1 and BRCA2 variants in a large, unselected Chinese Han breast cancer cohort. J Hematol Oncol. 2021;14:18.
Lefter M, Vis JK, Vermaat M, den Dunnen JT, Taschner PEM, Laros JFJ. Mutalyzer 2: next generation HGVS nomenclature checker. Bioinformatics. 2021;37:2811–7.
Chen S, Francioli LC, Goodrich JK, Collins RL, Kanai M, Wang Q, et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature. 2024;625:92–100.
Walker LC, Hoya M, Wiggins GAR, Lindy A, Vincent LM, Parsons MT, et al. Using the ACMG/AMP framework to capture evidence related to predicted and observed impact on splicing: Recommendations from the ClinGen SVI Splicing Subgroup. Am J Hum Genet. 2023;110:1046–67.
Tavtigian SV, Harrison SM, Boucher KM, Biesecker LG. Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines. Hum Mut. 2020;41:1734–7.
Makhnoon S, Davidson E, Shirts B, Arun B, Shete S. Practices and Views of US Oncologists and Genetic Counselors Regarding Patient Recontact After Variant Reclassification: Results of a Nationwide Survey. JCO Precis Oncol. 2023;7:e2300079.
Tudini E, Andrews J, Lawrence DM, King-Smith SL, Baker N, Baxter L, et al. Shariant platform: Enabling evidence sharing across Australian clinical genetic-testing laboratories to support variant interpretation. Am J Hum Genet. 2022;109:1960–73.
Li L, Tian X, Woodzell V, Gibbs RA, Yuan B, Venner E. Tracking updates in clinical databases increases efficiency for variant reanalysis. Genet Med Open. 2024;2:101841.
Fortuno C, Lee K, Olivier M, Pesaran T, Mai PL, de, et al. Specifications of the ACMG/AMP variant interpretation guidelines for germline TP53 variants. Hum Mutat. 2021;42:223–36.
Mester JL, Ghosh R, Pesaran T, Huether R, Karam R, Hruska KS, et al. Gene-specific criteria for PTEN variant curation: Recommendations from the ClinGen PTEN Expert Panel. Hum Mutat. 2018;39:1581–92.
Luo X, Maciaszek JL, Thompson BA, Leong HS, Dixon K, Sousa S, et al. Optimising clinical care through CDH1-specific germline variant curation: improvement of clinical assertions and updated curation guidelines. J Med Genet. 2023;60:568–75.
Tuffaha HW, Mitchell A, Ward RL, Connelly L, Butler JRG, Norris S, et al. Cost-effectiveness analysis of germ-line BRCA testing in women with breast cancer and cascade testing in family members of mutation carriers. Genet Med. 2018;20:985–94.
Koldehoff A, Danner M, Civello D, Rhiem K, Stock S, Müller D. Cost-Effectiveness of Targeted Genetic Testing for Breast and Ovarian Cancer: A Systematic Review. Value Health. 2021;24:303–12.
Sun L, Brentnall A, Patel S, Buist DSM, Bowles EJA, Evans DGR, et al. A Cost-effectiveness Analysis of Multigene Testing for All Patients With Breast Cancer. JAMA Oncol. 2019;5:1718–30.
Davidson AL, Kondrashova O, Leonard C, Wood S, Tudini E, Hollway GE, et al. Analysis of hereditary cancer gene variant classifications from ClinVar indicates a need for regular reassessment of clinical assertions. Hum Mutat. 2022;43:2054–62.
Walsh N, Cooper A, Dockery A, O’Byrne JJ. Variant reclassification and clinical implications. J Med Genet. 2024;61:207–11.
Loong L, Garrett A, Allen S, Choi S, Durkie M, Callaway A, et al. Reclassification of clinically-detected sequence variants: Framework for genetic clinicians and clinical scientists by CanVIG-UK (Cancer Variant Interpretation Group UK). Genet Med. 2022;24:1867–77.
Funding
CF was supported by a grant from the National Breast Cancer Foundation, Australia (IIRS-21-102). ABS and ALD were supported by NHMRC funding (APP177524). Collation of VUS through ICCon was supported by funding from the Peter MacCallum Cancer Foundation. Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Contributions
CF assisted with methodology, analysed data and wrote the first draft of the manuscript. EJP assisted with data acquisition and coordination. ALD, JH, GI, MEM, and MP assisted with data analysis. AMC, AD, VF, MJF, HGMF, CBN, NKP, LW, and RW provided data and/or answered the survey. VB, CE, AJ, MM, VSK, RS, and MW answered or assisted with answering the survey. HS assisted with methodology and answered the survey. PAJ assisted with methodology, answered the survey, supervised CF and critically reviewed the manuscript. ABS conceived the project and acquired funding, designed the methodology, supervised CF and critically reviewed the manuscript. All authors read and provided input for the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This project has been approved by the QIMR Berghofer Medical Research Institute Human Research Ethics Committee (HREC) under QIMR HREC Approval P1051.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41431_2024_1705_MOESM2_ESM.xlsx
Suggested clinically-relevant classifications from application of gene-specific ACMG/AMP criteria and points system (a-e)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fortuno, C., Cops, E.J., Davidson, A.L. et al. Unrecognised actionability for breast cancer risk variants identified in a national-level review of Australian familial cancer centres. Eur J Hum Genet 32, 1632–1639 (2024). https://doi.org/10.1038/s41431-024-01705-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-024-01705-9