Abstract
Protein methylation is a functionally important post-translational modification that occurs on diverse amino acid residues. The current proteomics approaches are inefficient to discover the methylation on residues other than Arg and Lys, which hinders the deep understanding of the functional role of rare protein methylation. Herein, we present a methyl-specific metabolic labeling approach for global methylome mapping, which enable the acquisition of methylome dataset covering diverse methylation types. Interestingly, of the identified methylation events, His methylation is found to be preferably occurred in C3H1 zinc fingers (ZFs). These His methylation events are determined to be Nπ specific and catalyzed by CARNMT1. The His methylation is found to stabilize the structure of ZFs. U2AF1 is used as a proof-of-concept to highlight the functional importance of His methylation in ZFs in RNA binding and RNA metabolism. The results of this study enable novel understanding of how protein methylation regulates cellular processes.
Similar content being viewed by others
Introduction
Protein methylation catalyzed by SAM-dependent methyltransferase1,2,3 represents a major type of post-translational modification (PTM) involved in various biological processes. Methylation on Lys (K) and Arg (R) residues have been extensively studied and are acknowledged to play prominent roles in the regulation of diverse biological processes including epigenetic regulation of gene transcription, RNA processing, DNA damage repair, and signal transduction4,5,6,7,8,9. Compared with the widely studied methylation at Arg and Lys residues, the methylation at other residues were largely overlooked. Currently, there are vast number of known Arg methylation events (11246 events) and Lys methylation events (5314 events) documented in the PhosphoSitePlus database10 (https://www.phosphosite.org/), in contrast to the annotation of only 14 sites for methylation at residues other than Arg and Lys in humans. Recent studies indicated that the methylation on residues, including Cys (C) methylation11, Glu (Q) methylation12 and His (H) methylation13,14, are also function important and play important roles in the regulation of diverse biological processes.
Methylation can occur at 8 amino acid residues to produce up to 11 types of methylation forms (Fig. 1a and Supplementary Table 1), including 3 forms on Lys (Mono-me-Lys, Di-me-Lys and Tri-me-Lys) and 2 forms on Arg (MMA and DMA). To dissect the functional landscape of protein methylation, especially the rare forms, it is necessary to analyze all methylation forms simultaneously. Unfortunately, identification of methylation sites with high confidence is difficult because the mass change introduced by methyl group is identical to some amino acid substitutions (Supplementary Table 2), which is particularly complicated when the amino acid substitution occurs close to the putative methylation sites. To improve the confidence of methylation identification, hM-SILAC (heavy methyl stable isotope labeling by amino acids in cell culture) strategy15 was usually used to distinguish the genuine methylation from potential false positives based on the presence of heavy and light methyl pairs. However, when all 11 methylation types on 8 amino acid residues are considered, 26 various modifications (Supplementary Table 3) need to be considered. The large number of possible methylation forms result in an unmanageable number of combinations for database search algorithms to match. Moreover, the methionine used to label the methylation was also incorporated into proteins, which resulted in methionine containing peptide pairs in the LC-MS/MS analysis16. Therefore, the methylated peptides could not be differentiated from the methionine containing peptides during the precursor scans. For these reasons, the systematical survey into the global landscape of protein methylation, especially the rare methylation forms, is still a big challenge.
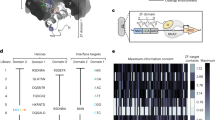
a Amino acid residues that can be methylated. b Workflow: Cells are differentially grown in light and methyl specific heavy Met, and are mixed at the ratio of 1:1 and lysed together. Proteins are digested with trypsin and analyzed by mass spectrometry. The methylated peptides are differentiated from methionine containing peptides by their different mass difference. By utilizing the correlation between the isotopic mass difference and the number of introduced methyl groups, the virtual MS/MS spectra corresponding to the unmethylated peptides are deduced by removing the mass of methylation modification in precursor mass and the MS2 peaks respectively. Then the extracted spectra are used for the methylation identification. c Example spectra for the in-silico demethylation approach. The twin peaks with identical mass were the peaks without methylation and the mass were kept, the twin peaks with a mass difference of 3*N were the peaks with methyl and the mass were kept after removing the mass of methyl. The deduced peaks were compared with the b or y ions for the identified sequence, showing that the ions without methyl can be deduced from the twin peaks with a mass difference of 3*N. d Methylated events identified in HEK293 cells and HepG2 cells. e The His methylation occurs exclusively on C3H1 ZF domains. Our Data: The information of total ZF proteins were obtained by subjecting our MS data to the traditional database searching. Deposited data: MS files of individually over-expressed ZF proteins were downloaded from the public database and analyzed. f Summary of identified methylated His peptides from C3H1 ZFs.
In this work, we presented a dedicated methyl-specific metabolic labeling strategy to allow the simultaneous identification of 11 methylation types on 8 amino acid residues. We found that His is the 3rd most extensively methylated residues. Interesting, among 13 identified His methylation sites, 5 were observed on the His residues of C3H1 ZF ___domain. We further confirmed the structure of the His methylation in C3H1 ZF domains to be Nπ specific and validated the CARNMT1, a methyltransferase previously known to methylate only small molecules17,18, to be the methyltransferase responsible for the His methylation at C3H1 ZFs. Molecular stimulation analysis revealed that the methylated His interact with a Tyr (in U2AF1) in ZF by forming π-π stacking and the methylation can stabilize the overall ZF ___domain. We also clarified that the structure stabilization by His methylation is a general mechanism in methylated ZFs. Therefore, our results deciphered a novel and general mechanism underlying the requirement of His methylation for maintaining the proper structure and function of some specific C3H1 ZFs.
Results
Methyl-specific metabolic labeling allows the global analysis of diverse methylation forms
To allow global analysis of methylproteome, we developed a new methyl-specific metabolic labeling approach using the mass shift introduced by the metabolic labeling with a special heavy methionine (-CD3, and 13COOH (Supplementary Fig. 1)). In contrast to the traditional hM-SILAC, this new labeling strategy can differentiate the methylated peptides (~3 Da per methyl) from methionine containing peptides (~4 Da per methionine) (Fig. 1b). Based on this, the number of methyl groups in MS1 and MS2 fragments are determined to deduce a virtual MS/MS spectrum with fragment ions carrying no methyl group (Fig. 1b, c), i.e. in-silico demethylated MS/MS spectra, and the peptide sequences could be identified without setting methylation as variable modifications, which dramatically reduce searching space. Subsequently, the methylation sites are located by considering the fragment ions carrying the methyl groups in the original heavy and light spectra. As no pre-setting of the methylation form is required, this approach enables the simultaneous analysis of 11 methylation forms on 8 residues.
Prior to methylome analysis, the labeling efficiency of the metabolic labeling was investigated and high labeling efficiency (around 99%) was observed (Supplementary Table 5), which indicated the high SILAC incorporation efficiency. The proposed strategy was applied to analyze the methylome of HEK293 cells and HepG2 cells, overall, 594 methylation forms on 498 methylated sites from 215 proteins were identified (see Supplementary Data 1 and Supplementary notes). Methylation was identified on all 8 known methylated residues (Fig. 1d), indicating the robustness of this approach in global methylome mapping. As the methylated Arg sites have obvious Gly-Arg-rich consensus sequences19, we examined the sequence patterns of the methylated Arg sites identified in this study and the consensus sequences were observed (Supplementary Fig. 2). The high consistence of the sequence pattern of the identified Arg methylation sites with known pattern suggests these sites were of high confidence. The obtained methylome dataset covers diverse methylation types and was thus used to evaluate the relative occurrence frequency of different methylation types. As can be seen in Fig. 1d, Arg is the most frequently modified residue and Lys follows. Much fewer methylation events were identified on other residues including His, Asp, Asn, Glu, Gln and Cys.
As shown in Fig. 1d and Supplementary Fig. 3, His is the 3rd most extensively methylated residues in our dataset. Totally, we identified 13 methylated His sites on 12 proteins. Of these 13 sites, 5 His methylation sites were identified in ZF ___domain of 5 ZF proteins (Fig. 1f and Supplementary Table 6). The methylation of H22 in NUPL2 was observed in HEK293 cells, and that of H37 in U2AF1 was identified in HepG2 cells. Other 3 methylation sites, H215 in ZC3H8, H242 in ZC3H18 and H123 in ZC3H15 were observed in both cell lines. To confirm the presence of His methylation, above-mentioned proteins with flag tag were expressed in HEK293 cells. All the over-expressed proteins were purified and subjected to LC-MS/MS analysis for the identification of methylation sites. All 5 methylation sites were identified with high confidence and an additional site was identified for ZC3H8 (Supplementary Table 7), indicating the His of ZF domains in these proteins are indeed methylated.
The His methylation mainly occurs on C3H1 ZF domains
Among the 3 types of ZF domains (Supplementary Fig. 4), only 2, i.e. C3H1 and C2H2, have His residues20,21. According to the Uniprot database (https://www.uniprot.org/), 973 and 58 proteins are annotated to have C2H2 and C3H1 ZF domains for human beings, respectively. It’s very interesting that all the 5 His methylation sites were only observed on C3H1 ZF ___domain even though the presence of at least 10 times more C2H2 ZF proteins in human proteome. To confirm the exclusive occurrence of His methylation on C3H1 ZF proteins rather than C2H2 ZF proteins, we analyzed the deposited raw MS data of the purified recombinant proteins reported by Gygi et al.22,23. In their dataset, the MS data of 23 C3H1 ZF proteins and 94 C2H2 ZF proteins (see Supplementary Data 2 for the gene lists) were available. Of the 23 C3H1 ZF proteins, 7 His methylation sites were identified in the ZF His (Supplementary Table 7), however, no such methylation was identified for the C2H2 ZF His even though much more proteins were analyzed (Fig. 1e). This is consistent with our proteomics analysis that the His methylation was exclusively observed on C3H1 ZF ___domain. To investigate the stoichiometry of these methylation sites, our proteomics data were also subjected to the traditional database searching24 to identify corresponding unmodified form, however, the unmodified forms were not identified, indicating the extremely low level of the unmodified forms. Additionally, in the proteins from our over-expressed samples as well as in the Gygi’s MS data of recombinant proteins, higher intensity was mostly found for the modified form rather than the unmodified form (Fig.1f), further indicating the high stoichiometry of these His methylation in C3H1 ZF domains. While stoichiometry itself is not an absolute indicator of functional modification sites, high stoichiometry sites are unlikely to occur spuriously and are therefore more likely to have a function. For protein methylation, it requires an energy demand of 12 molecules of ATP per methylation event25 and is highly energy costing. If the cell uses such high energy cost to modify a site to high stoichiometry, this site is more likely to have a functional role. Therefore, His methylation has the potential to affect the function of a large pool of C3H1 ZF proteins.
Nπ of His on C3H1 ZF domains is methylated
As shown in Fig. 2a, in the side chain of His, two nitrogens can be modified13, namely Nτ-me and Nπ-me. Therefore, it’s important to determine the precise modification position. Totally, we identified 6 methylated peptides from the C3H1 ZF domains. We synthesized all the 6 peptides with either Nτ-me or Nπ-me form. In proteomics studies, co-elution of modified peptides during chromatographic separation is used as golden standard to differentiate the modification structure26,27. In this study, two different chromatography techniques were applied (Fig. 2b). Firstly, as can be seen in Supplementary Fig. 5, methylation on different nitrogen can cause difference in hydrophobicity and therefore resulting in different retention in the RPLC separation. The in-vivo states for 5 sites in proteins (ZC3H8, ZC3H15, ZC3H18 and U2AF1) were found to co-eluted with the Nπ methylated form rather than the Nτ methylated form, indicating the modification forms on these His were Nπ-me form (Fig. 2c and Supplementary Fig. 6). Moreover, as methylation may also affect chelating ability of these His residues, a RPLC independent approach was utilized to confirm the modification structure. Compared to the Nτ-me-His counterparts, the Nπ-me-His containing peptides bind to the Cu-IMAC matrix with slightly higher affinity and were eluted with higher imidazole concentration. And the methylated peptides of the in-vivo states for 4 sites in 3 proteins (ZC3H8, ZC3H15 and ZC3H18) co-eluted with the synthesized Nπ-me-His counterpart (Fig. 2d and Supplementary Fig. 7), further indicating the methylation on these methylation sites belongs to the Nπ-me-His forms. Due to the lower available sample amount and the sample loss in the two desalting processes during the Cu-IMAC analysis, the peptides for U2AF1 cannot be identified in the MS analysis. Besides, due to the extremely low available amount of NUPL2, this protein was not subjected to methylation structure determination. As summarized in Fig. 2e, all confirmed His methylation sites in C3H1 ZFs are Nπ methylation.
a Nitrogen positions that can be modified by methylation on His residue. b Workflow for determining the modification structure. c In-vivo methylated peptides (FDH*DAEIEK) co-eluted with the synthesized Nπ methylated peptides and eluted slightly earlier than the Nτ methylated peptides in the RP-LC separation. d In-vivo methylated peptides (FDH*DAEIEK) co-eluted with the synthesized Nπ methylated peptides in the Cu-IMAC separation. e Summarized results for the structure determination.
His methylation stabilizes the structure of C3H1 ZFs
To reveal the functional role of His methylation in regulating the C3H1 ZFs, we focused on U2AF1, a protein with available C3H1 ZF structure28,29. Hence, the molecular dynamics simulations were performed on un-methylated and methylated (H37) U2AF1/RNA complexes to analyze the influence of His methylation on the conformation of the ZF. The root-mean-square deviations (RMSD) of the Cα atom and the radius of gyration (Rg) were used to characterize the global dynamic state of the ZF. Compared with the protein with higher values of RMSD and RMSF, the protein with lower RMSF and RMSD values exhibit smaller fluctuations during the molecular simulations, indicating that their thermodynamic properties are more stable. As shown in Fig. 3, the occurrence of His methylation resulted in a decrease in RMSD (Fig. 3a) and Rg (Fig. 3b), indicating that the ZF became more stable in thermodynamic properties after His methylation. Furthermore, the results of root mean square fluctuation (RMSF) indicated that the methylation modification significantly reduced the fluctuation of each residue of the ZF (Supplementary Fig. 8). In detail, the side chain of H37 exhibited high flexibility without methylation (Fig. 3c), which was reflected in the two different relative positions of the imidazolyl group of H37 and the aromatic ring of Y21 (parallel or vertical). However, the introduction of methyl group resulted in the parallel position for H37 and Y21 during the simulations (Fig. 3c), suggesting that His methylation was able to stabilize the π-π stacking interaction between H37 with Y21. Further analysis found that the increase in the stability of the ZF shortened the distance between S34:OG and the imidazolyl of H29, which might be able to facilitate the recognition of RNA by S34 and H29. Those results indicated that the structure alteration of ZF caused by His methylation, especially for the π-π stacking between H37and Y21, might regulate the function of U2AF1 through affecting the recognition of pre-mRNAs by U2AF1.
a Comparison of the calculated RMSD between ZF of unmethylated and methylated U2AF1. b Comparison of the calculated Rg between unmethylated and methylated U2AF1. c The influence of methylation (His37) on ZF. A–C Comparison of the ZF structure in unmethylated U2AF1 (A, B) and methylated U2AF1 (C). The key residues that bond to Zn2+ were highlighted by salmon sticks and the Zn2+ was showed as gray spheres. The S34 and H29 were highlighted by violet purple sticks, while the Y21 was colored by green. D–F Distribution frequency of the distance between H37:ND1 and C27:O in Un-methylated and Methylated U2AF1(D), the dihedral of H37:CA-CB-CG-CD2 in Un-methylated and Methylated U2AF1(E), the distance between S34:OG and imidazole of H29 in Un-methylated and Methylated U2AF1(F). d Comparison of the sequences and structures between the ZFs that could be methylated and the ZFs that cannot be methylated. The residues, formed interactions with Zn2+, were highlighted with black boxes in the sequences.
With the aim to clarify whether the structural influence caused by His methylation is a general mechanism for specific C3H1 ZFs, we carried out molecular dynamics simulations on other C3H1 ZF structures which have been detected to undergo His methylation (Supplementary Data 3 and Supplementary Data 5), as well as the C3H1 ZF structures that cannot be methylated (Supplementary Data 3). We divided these C3H1 ZFs into three classes (Fig. 3d): 1) C3H1 ZFs with methylated His that form the π-π stacking interactions with aromatic residue; 2) C3H1 ZFs with unmethylated His that can form the π-π stacking interactions with aromatic residue; 3) C3H1 ZFs with unmethylated His that is unable to form the π-π stacking interactions with aromatic residue. For the first class C3H1 ZFs, the occurrence of methylation was able to stabilize the π-π stacking interaction of His and aromatics by maintaining the imidazole group of His and the aromatic ring of aromatic residue in the parallel position. Consistently, the introduction of methyl group reduced the RMSD values of first class C3H1 ZFs (except ZC3H8_246-271), indicating an increase in the stability of the structure of C3H1 ZFs (Supplementary Fig. 9 and Supplementary Fig. 10). On the contrary, this phenomenon was not detected in the second and third classes C3H1 ZFs (Supplementary Figs. 11–14). Those results indicated that for the C3H1 ZFs that can be methylated, the His methylation was able to enhance the stability of C3H1 ZF structure by stabilizing the π-π stacking interaction of His and aromatics.
CARNMT1 interacts with and modify C3H1 ZF proteins in vivo and in vitro
We next sought to dissect how His methylation on C3H1 ZF proteins affect the function of ZF proteins. To answer this question, we need to determine the enzyme responsible for this modification. Among histidine methyltransferase, we noted that the modification preference of CARNMT1 on carnosine is Nπ specific18, which is in accordance with the methylation structure on ZF proteins determined in this study. Therefore, we speculated that CARNMT1 might be responsible for the methylation on the identified C3H1 proteins. We first examined whether CARMMT1 interacts with these ZF proteins. For this, we co-expressed Flag-ZF proteins and HA-CARNMT1 protein in HeLa cells and conducted immunoprecipitations with Flag antibody. Western blot showed that HA-CARNMT1 was apparently co-precipitated (Fig. 4a). Additionally, the reverse immunoprecipitations with HA antibody also confirmed these interactions (Supplementary Fig. 15).
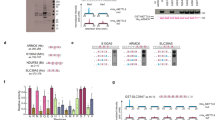
a The interaction between CARNMT1 and ZF proteins was confirmed by the co-purification of the HA tagged CARNMT1 with the Flag tagged ZF proteins. 2 times each experiment was repeated independently with similar results. b Deletion of CARNMT1 in HeLa cells. 2 times each experiment was repeated independently with similar results. c His methylation of over-expressed U2AF1 cannot be observed after the deletion of CARNMT1. d Supplementation of wild-type and mutated CARNMT1 in CARNMT1-KO cells. 2 times each experiment was repeated independently with similar results. e The methylation of over-expressed U2AF1 in CARNMT1 KO cells can be recovered by the supplementation of WT CARNMT1 and was not recovered by the supplementation of mutated CARNMT1. f ZF ___domain peptides corresponding to ZF1 of U2AF1 can be methylated by CARNMT1 in the in-vitro methylation assay. g ZF1 in the purified U2AF1 from E.coli can be methylated by CARNMT1 in the in-vitro methylation assay, while the ZF2 cannot be modified. The retention time information can be found in the identification result in Supplementary Table 8.
To validate the enzyme-substrate relationship between CARNMT1 and these C3H1 ZF proteins in vivo and examine whether CARNMT1 is the major methyltransferase for the ZF proteins, we generated CARNMT1 knock-out cell lines using CRISPR/Cas9-mediated gene editing30 (Fig. 4b and Supplementary Fig. 16) and analyzed the corresponding methylation level changes on exogenously expressed ZF proteins. Interestingly, when we over-expressed these C3H1 ZF proteins in the CARNMT1 KO and WT HeLa cells (Fig. 4c and Supplementary Fig. 17), the methylation was only detected in ZF proteins over-expressed in WT cells, while the methylation disappeared completely in over-expressed proteins from the KO cells for all tested 5 sites in 4 proteins (U2AF1, ZC3H8, ZC3H15 and ZC3H18), indicating that the CARNMT1 is indeed responsible for the His methylation on C3H1 ZFs, at least for the sites identified in this study. To further examine this, we tested whether the methylation for U2AF1 in KO samples can be recovered by the supplementation of CARNMT1 (Fig. 4d). Importantly, the methylation of U2AF1 was observed in CARNMT1 KO cells supplemented with wild type CARNMT1 but cannot be recovered by the supplementation of methylase-dead-mutated CARNMT118 (Fig. 4e), providing further evidence for the role of CARNMT1 in conducting His methylation in C3H1 ZF proteins.
To examine whether CARNMT1 methylate C3H1 ZFs in-vitro, CARNMT1 was expressed and purified from E. coli, followed by incubation with the synthesized ZF domains. Interestingly, all tested ZF domains can be modified with a mass shift of +14, corresponding to the methylation modification (Fig. 4f and Supplementary Fig. 18). To confirm the methylated residue, we used the ZF ___domain peptides that were already methylated (Nτ-me or Nπ-me) in the corresponding identified His methylation sites. When the modified ZF domains were incubated with the recombinant CARNMT1, additional +14 mass shifts were not observed (Supplementary Fig. 18). These data indicate that the methylation occurs on the site identified in our proteomics approach. Interestingly, the Nτ methylated ZF domains cannot be modified either, possibly due to the importance of this position for the His binding in the CARNMT1 pocket, as in the case for modifying carnosine18. We next examined whether ZF proteins can be methylated by CARNMT1. For this, U2AF1 was purified from E.coli (Supplementary Fig. 19) and subjected to the in-vitro methylation assay. As can be seen in Fig. 4g, consistent with the proteomic result, the His in ZF1, but not that in ZF2 of purified U2AF1 was found to be modified by CARNMT1, further confirming the catalytic specificity of CARNMT1 in His methylation.
Evidence that His methylation regulates splicing activity of U2AF1
His methylation could induce conformation change of the methylated ZFs, thus might regulate function of ZF proteins. To test this, we focused on U2AF1, a core splicing factor that participates in the regulation of more than 80% of all alternative splicing events (ASEs)31. We first examined the global influence of the CARNMT1 KO in pre-mRNA splicing using RNA sequencing (RNA-seq). As shown in Fig. 5a, the deletion of CARNMT1 resulted in 3026 altered ASEs, with the most significantly changed ASEs exhibiting the skipped exons (SEs) (representing 74.6% of altered ASEs). These results suggest an important role of CARNMT1 in regulating alternative splicing.
a Distribution of changed splicing events when CARNMT1 is deleted, blue for down-regulated and red for up-regulated. b Distribution of changed splicing events when U2AF1 is knocked down, blue for down-regulated and red for up-regulated. c Recovery experiments by supplementing either wild-type or mutated CARNMT1 into CARNMT1 KO cells. GST: Flag-GST, WT: Flag-CARNMT1, M1: Flag-CARNMT1 (D316A), M2: Flag-CARNMT1 (F313A/D316A). 2 times each experiment was repeated independently with similar results. d Two representative altered splicing events were subjected to recovery experiments and quantification of exon inclusion ratio. The data represent mean ± SD. P-value, one-tailed paired t-test. (n = 3 biological replicates). Source data are provided as a Source Data file. e Scheme for illustrating the binding between U2AF complex and pre-mRNA. f Volcano plot showing the alteration in inclusion level for the U2AF1 regulated splicing events (SE events grouped by the nucleotide ahead AG sites) when CARNMT1 is deleted. Source data are provided as a Source Data file. g Altered splicing events of mini-gene expressed in either CARNMT1 WT or KO cells and quantification of exon inclusion ratio. The data represent mean ± SD. P-value, one-tailed paired t-test. (n = 3 biological replicates). Source data are provided as a Source Data file. h eCLIP result for the RNA binding landscape of U2AF1 in CARNMT1 KO and WT cells. i)The binding free energy between U2AF1 and RNA. j MST results (EC50) for the measurement of in-vitro binding between U2AF1 and RNAs. k Structure for illustrating the binding between ZF of U2AF1 and 3’ss of pre-mRNA.
To understand how these ASE altered in CARNMT1 KO might reflect the effect of loss of His methylation of U2AF1, we also conducted RNA-seq for U2AF1 KD cells. Consistent with the role of U2AF1 as a core splicing factor, a large number of splicing events (25906) were significantly altered (Fig. 5b). We observed a dominant trend favoring exon exclusion in SE and increased intron retention (RI), which differs from CARNMT1 KO cells (Fig. 5a). This result suggests the regulation of CARNMT1-mediated His methylation in RNA splicing is more complex than solely through U2AF1. To understand how His methylation of U2AF1 affect its functions, we next specifically focused on splicing events commonly regulated by U2AF1 and CARNMT1. First, to clarify whether the altered splicing regulation was associated with the methyl-transfer activity of CARNMT1, we selected two events and examined their splicing by re-expressing either the wild-type or methylase-dead-mutant CARNMT1 into the CARNMT1 KO cells (Fig. 5c). As expected, wild-type but not the mutant CARNMT1 rescued the splicing changes (Fig. 5d), suggesting that the observed splicing alterations were indeed caused by the methyl-transfer function of CARNMT1.
Given the key role of U2AF1 in the recognition of 3’SS, we next focused on the 3’ splicing sites (3’ SS) of the altered SEs commonly regulated by CARNMT1 and U2AF1. When the 3’SS was grouped by the preceding nucleotide at AG dinucleotide (Fig. 5e), more notable decline of inclusion levels was observed for the 3’SS containing TAG (Fig. 5f), suggesting that U2AF1 His methylation might be especially important for its role at the TAG 3’SS. To validate the altered splicing regulation in CARNMT1 KO cells and to examine how methylation of U2AF1 affects its role in different type of 3’SS, we performed splicing assays using the mini-gene of ATG16L1 containing the alternative exons affected by both CARNMT1 and U2AF1. Consistent with the results from endogenous RNAs, the inclusion level of the alternative exon in the mini-gene was also decreased. However, the decrease was also observed when the 3’SS in the mini-gene was mutated from TAG to CAG or AAG (Fig. 5g). We speculate that the bias on TAG on endogenous RNAs could be due to a context-dependent effect. Collectively, our results indicate that CARNMT1 regulates the pre-mRNA splicing, partially through His methylation on U2AF1.
His methylation increases the binding ability of U2AF1 at 3’ splicing sites
To further examine how His methylation modulates U2AF1-mediated splicing regulation and affects U2AF1’s recognition of 3’ splicing sites, we dissected transcriptome-wide U2AF1-RNA interactions with or without His methylation. For this, enhanced crosslinking and immunoprecipitation (eCLIP)32 was performed for U2AF1 in CARNMT1 KO and WT cells. Significantly, upon deletion of CARNMT1, obviously weakened binding signal of U2AF1 was observed at AG-dinucleotide (Fig. 5h), indicating His methylation increases the binding ability of U2AF1 at 3’ splicing sites. We noted an increase in the signal at poly-pyrimidine region, which could be due to compensation of increased binding of U2AF2 at these sites. When we compared the read distribution at different regions of pre-mRNAs, we did not detect significant difference between CARNMT1 KO and WT cells (Supplementary Fig. 20), indicating the His methylation does not significantly affect the distribution of U2AF1 along pre-mRNAs.
Consistent with the eCLIP data, the data of MM/GBSA obtained by molecular stimulation analysis also showed that His methylation increase the binding between U2AF1 and various 3’ splicing sites (TAG/CAG/AAG) (Fig. 5i). To further validate these findings in-vitro, using purified U2AF1 protein with or without methylation by purified CARNMT1, we performed the MicroScale Thermophoresis (MST)33 experiment to measure the binding affinity for 3 synthesized RNA oligos carrying different 3’SS corresponding to TAG, CAG or AAG, respectively. Consistent with our eCLIP and mini-gene data, increased binding affinity was observed in all 3 tested RNAs for methylated U2AF1 (Fig. 5j and Supplementary Fig. 21). To examine whether this is also true for U2AF1’s first ZF ___domain (18-37aa), the binding affinity between synthesized RNAs and synthesized ZF ___domain with or without methylation was also evaluated. Indeed, increased binding affinity was observed for methylated first ZF ___domain (Fig. 5j and Supplementary Fig. 22). All above data indicated that His methylation can affect U2AF1’s recognition of 3’ splicing sites by increasing the binding ability at AG position (Fig. 5k).
Overall, our data indicates that His methylation can stabilize the conformation of certain C3H1 ZFs and the introduced structure change can indeed affect the RNA binding and the subsequent RNA processing.
Discussion
To enable the identification of methylation on multiple residues, we presented a new methyl-specific metabolic labeling approach, which can differentiate the methylated peptides (~3 Da per methyl) from methionine containing peptides (~4 Da per methionine), and thereby significantly reduce the search space. Metabolic labeling using heavy form of methionine is a widely utilized approach to improve the confidence of methylpeptide identification in the analysis of protein methylation15. However, due to the limitation of SILAC labeling, the tissue samples are currently not suitable to be labeled and analyzed by our strategy. Besides, as no specific enrichment was conducted, the signal of methylated peptides might be suppressed by the abundant unmethylated peptides and depth of the methylome analysis was restricted, which might miss the identification of some methylated peptides of low abundance. Regardless, our approach could work as an important tool to uncover a wealth of information about protein methylation, especially in studies where multiple methylation forms need to be analyzed.
This approach was applied to analyze the methylome of HEK293 cells and HepG2 cells using sample preparation scheme with SCX fractionation, which yielded the identification of 594 methylation forms on 498 methylated sites of 215 proteins. As expected, Arg and Lys are the most frequently modified residue. Much fewer methylation events were identified on other residues including His, Asp, Asn, Glu, Gln and Cys. The amino acid residue preference of protein methylation largely followed this order: Arg≫Lys>His>Glu≈Gln≈Asp≈Asn. The identified methylated Arg sites have obvious Gly-Arg-rich consensus sequences, indicating high confidence of the identified methylation sites. However, due to the extremely low abundance of non-Lys/Arg methylation, further experiments are required to confirm the reliability of the identified rare methylation forms. Among the 594 identified methylation forms, only 13 (2.2%) were observed on His. Interestingly, of these 13 sites, 5 His methylation sites were identified in C3H1 ZF ___domain of 5 ZF proteins. The methylome of HEK293 cells was also analyzed by this approach using another sample preparation scheme with hRP fractionation34, which resulted in the identification of 294 methylation sites (as detailed in the Supplementary notes). Similarly, of the 5 identified His methylation sites, 2 sites were observed in the C3H1 ZF ___domain of ZC3H15 and ZC3H18, and the methylation sites (H242 in ZC3H18 and H123 in ZC3H15) are consistent with those identified in the methylome analysis using sample preparation scheme with SCX fractionation. The His methylation sites identified C3H1 ZFs were validated by over-expressing the corresponding ZF proteins with flag tag that can be purified. And the His methylation sites were unambiguously observed in the over-expressed and purified proteins, confirming that all the 5 His methylation sites are correct identifications. Additionally, we confirmed the structure of the His methylation in these C3H1 domains to be Nπ specific by two independent approaches.
To dissect the functional significance of His methylation at C3H1 ZF proteins, we utilized U2AF1 as proof-of-concept. To investigate whether the changed binding affinity caused by His methylation could affect the splicing activity of U2AF1, we conducted in-depth RNA sequencing for CARNMT1-knockout and U2AF1-knockdown cells and found the His methylation of U2AF1 affect the proper splicing of extensive alternative splicing events, suggesting the His methylation on U2AF1 is essential for its proper splicing function. Above results were also confirmed by a recent study35 reported by Shinkai’s group that investigated the biological functions of CARNMT1. Shinkai’s study also noticed that the absence of His methylation can lead to impaired alternative splicing performance of U2AF1, and this is consistent with the result of our study. CARNMT1 is a rarely studied methyltransferase, however, the results of our study and Shinkai’s study, presented two independent confirmations of the critical role of CARNMT1 in regulating RNA processing (Supplementary Fig. 23) and this may open-up a new field for further investigation.
The methyl-specific metabolic labeling approach presented in this study enables the identification of endogenous His methylation on C3H1 ZF proteins for the first time. These His methylation sites were of high stoichiometry and were functionally important. In the human proteome, there are around 160 C3H1 ZFs at 58 proteins. However, only certain ZF are methylated. When analyzing the sequence characteristics of these methylated ZFs, as shown in Supplementary Fig. 24, consensus motif pattern was observed for the methylated ZFs. Additionally, in our study, we clarified the functional necessity of His methylation to the methylated ZFs, and revealed that the underlying mechanism is to regulate the conformation and structure stability of C3H1 ZFs. Additionally, we found that the absence of His methylation would result in reduced binding between the 3’ splicing sites and U2AF1, and this finding was systemically validated in-vivo by eCLIP experiment and in-vitro by MST analysis. Collectively, our study enables novel insights into the biological significance of His methylation in ZF proteins.
Although the underlying mechanism remains unclear, the biased effect on TAG was frequently found in the functional analysis of U2AF1, especially in the S34F mutation of U2AF129,36,37,38, suggesting the TAG splicing sites might be more sensitive to the alteration in U2AF1 binding ability. Splicing is a highly complex and sophisticatedly regulated process, involving hundreds of core splicing factors and regulators. For the recognition of 3’SS, in addition to the core AG sites, the poly-pyrimidine sequence, exonic splicing enhancer (ESE), exonic splicing silencer (ESS), intronic splicing enhancer (ISE), and intronic splicing silencer (ISS) of pre-mRNAs also make important contributions. Thus, the biased effect on TAG splicing sites caused by His methylation of U2AF1 might be a combined effect of all these factors.
C3H1 ZF and C2H2 ZF are two types of ZFs. The C3H1 ZF mainly involved in the RNA binding while the C2H2 ZF mainly involved in the DNA binding20. The RNA metabolism is a highly sophisticated process and RNA molecules have more diverse structures. Therefore, more structure diversity might be needed for the RNA binding proteins to function properly. Under certain circumstance, the His methylation might be required for the structure optimization to modulate the binding, as also exemplified in the methylation of Arg residues for regulating the RNA binding39, and this may be why the His methylation mainly occurs on C3H1 ZF domains. Therefore, our finding indicates the necessity of His methylation for regulating RNA processing.
Analyses of the TCGA database40 showed that the mRNA levels of CARNMT1 are much lower in KIRC (Kidney renal clear cell carcinoma) and KIRP (Kidney renal papillary cell carcinoma) than in normal kidney tissues (Supplementary Fig. 25). Additionally, the patients whose tumors with low levels of CARNMT1 had a much shorter survival duration than those whose tumors with high levels of CARNMT1 (Supplementary Fig. 25). This result supported the role of CARNMT1 in the clinical behavior of human KIRC/KIRP and revealed a relationship between CARNMT1 and clinical aggressiveness of KIRC/KIRP. The finding that the levels of CARNMT1 were positively correlated with longer survival in patients with KIRC/KIRP also underscores the potential to target the activity of CARNMT1 for the treatment of human cancer. However, because His residues on multiple proteins can be methylated by CARNMT1 and a small molecule of carnosine is also a target for the methylation by CARNMT118, it is important but difficult to build a direct link between the His methylation of individual substrate and the biological consequence. Therefore, further work is needed to clarify the broader effects of CARNMT1 at the cellular or tissue level.
In summary, we presented a methyl-specific metabolic labeling approach for global methylome mapping and found extensive His methylation at C3H1 ZFs. CARNMT1 was found to be the responsible methyltransferase and the His methylation is essential for the proper function of these methylated ZF proteins. Our study revealed the functional necessity of His methylation in ZF proteins and validated the underlying mechanism in-vivo by eCLIP experiment and in-vitro by MST analysis, which would certainly enable better understanding of how protein methylation regulates cellular processes.
Methods
Chemicals and reagents
Water used in the experiments was prepared by a Milli-Q system (Millipore). Formic acid (FA) was purchased from Fluka and acetonitrile (ACN, HPLC grade) from Merck. Daisogel ODS-AQ (5 μm, 12 nm pore) was purchased from DAISO Chemical CO., Ltd. Yeast extract, tryptone, agar, kanamycin and isopropyl β-D-thiogalactoside (IPTG) were obtained from Sangon Biotech. Fused-silica capillaries with 200 μm or 75 μm inner diameter were purchased from Polymicro Technologies. Lip2000 was from Thermo Fisher Scientific. Anti-CARNMT1 antibody (HPA026756) was bought from Sigma; Anti-U2AF1 antibody (60289-1-Ig), Anti-HA antibody (51064-2-AP) and Anti-FLAG antibody (20543-1-AP) was bought from Proteintech. Plasmids used in this study were custom purchased from geneppl. Peptides used in this study were custom purchased from scilight-peptide. All other chemicals and materials were purchased from Sigma-Aldrich.
hM-SILAC labeling and cell lysis for MS analysis
The light labeling media and heavy labeling media were prepared by adding L-Arginine, L-Lysine and either L-Methionine or L-Methionine-(carboxy-13C, methyl-D3) at a final concentration of 0.398 mM, 0.798 mM and 0.2 mM, respectively to the custom-purchased DMEM medium (Gibco) lacking L-Methionine, L-Arginine, and L-Lysine supplemented with 10% dialyzed fetal bovine serum (Gibco). HEK293 and HepG2 cells (ATCC) were grown at 37 °C in a humidified 5% CO2-containing atmosphere for 8 cell doublings in the light or heavy labeling medium. Cells were harvested by trypsin digestion and washed with PBS buffer (pH 7.4). Then the cells cultured under light methionine condition and heavy methionine condition were mixed at the ratio of 1:1 (cell number) and were resuspended in 10 volumes of ice-cold hypotonic lysis buffer (25 mM HEPES (pH 7.9), 5 mM KCl, 0.5 mM MgCl2 1 mM DTT, 1% v/v NP-40 and 1% v/v protease inhibitor cocktail). After being homogenized using a dounce homogenizer, the supernatant containing the cytoplasmic fraction was collected by centrifugation at 1000 g for 10 min. The debris was washed twice with hypotonic buffer and subsequently lysated by sonication in 10 volumes of ice-cold high salt lysis buffer (25 mM HEPES (pH 7.9), 350 mM NaCl, 0.5 mM MgCl2 1 mM DTT, 1% v/v NP-40 and 1% v/v protease inhibitor cocktail). After centrifugation at 25,000 g for 30 min, the nucleoplasmic fraction was collected. After being processed by acetone/ethanol precipitation, the precipitated proteins were resuspended in the denaturing buffer containing 8 M urea and 50 mM NH4HCO3 (pH 8.1) and the concentration of proteins was measured by the Bradford method. 2 mg of proteins from cytoplasmic fraction or nucleoplasmic fraction were reduced by 20 mM DTT at 37 °C for 2 h and alkylated by 40 mM iodoacetamide in the dark at room temperature for 40 min. After adding 50 mM NH4HCO3 (pH 8.1) to dilute the denaturing buffer, trypsin was added at an enzyme-to-protein ratio of 1:50 and was incubated for 16 h for digestion at 37 °C. The digestion was quenched by acidification with TFA to 1% (v/v) on ice and the resulting peptides were desalted with 60 mg HLB cartridge.
SCX fractionation
The desalted peptides were dissolved in loading buffer (7 mM KH2PO4 and 25% ACN, PH 2.8), and were loaded onto a 4.6 mm × 10 cm PL-SCX column (Agilient) connected to a HPLC system (HITACHI). Fractions were collected during a 40 min gradient from 5% to 35% solvent B (solvent A: 25%ACN, 7 mM phosphate buffer (pH 2.8); solvent B: the same as A with additional 500 mM KCl; flow rate: 1 ml/min). The frequency for fraction collection was as following: a fraction every 2 minute during 0–8 min and a fraction every 1 min during 8–40 min. All collected fractions were dried down to remove acetonitrile (ACN) by vacuum centrifugation. After removing salt using SPE column, the collected fractions were stored under −30 °C.
Nano-LC − MS/MS analysis
One tenth of sample from each elution fractions was subjected to the analysis by LC-MS/MS (n = 1 replicates). For the LC system, a nano-HPLC Dionex UltiMate 3000 (Thermo Scientific) is connected to a Q-Exactive mass spectrometer. The injected sample was first captured on a trapping column before being separated in an analytical column. Peptides are chromatographically separated by using a separation gradient of 180 min at a column flow rate of ~300 nL/min. The column effluent is directly introduced into the ESI source of the MS; HCD fragmentation is used on the Q-Exactive MS. The mass spectrometer was operated in a ‘Top 8’ data-dependent acquisition mode with dynamic exclusion enabled (30 s). Survey scans (mass range 300–1750 m/z) were acquired at a resolution of 70,000 at 200 m/z with the 8 most abundant multiply charged (z ≥ 2) ions selected with a 1.0 m/z isolation window for HCD fragmentation. MS/MS scans were acquired at a resolution of 70,000 at 200 m/z.
In silico demethylation of spectra for methyl peptide pair
Raw files were converted to mzML format by Proteome Discoverer (version 1.4.0.0). Peptide features were generated by using Dinosaur software, which is reported that around 98% of monoisotopic peaks can be accurately determined through this software41. Then, paired features were obtained by using the following criteria: 1) with the same charge state; 2) The mass difference between two features meet the requirement of equation MassH_Feature – MassL_Feature = 3.01883 × NMod + 4.022185 × NMet (Herein, MassH_Feature is the mass of heavy-feature (mass of the feature with larger mass, which is potentially the feature of heavy-labeled methylated peptide) and the MassL_Feature is the light-feature mass. NModϵ [1,6] is the range of the methyl count. NMet ϵ [0, 2] is the range of methionine count); 3) the retention time difference (RTH_Feature - RTL_Feature) between two features should be within the range of 10 ± 60 seconds; 4) the intensity ratio is required to be within the four folds. All paired features are supposed to come from the peptide with identical sequence but with different methyl labeling form, and the spectra corresponding to the un-methylated form are deduced from the spectra with paired features. Detailly, the precursor m/z of the spectrum corresponding to the un-methylated form is calculated using the equation MZPre = MZL_Pre–NMod×ML_Mod/ZPre or MZPre = MZH_Pre-(NMod× MH_Mod + 4.022185×NMet)/ZPre (MZL_Pre is the precursor m/z of the light-spectrum, which is the spectrum of light-feature. MZH_Pre is the precursor m/z of the heavy-spectrum. ML_Mod is the mass of light mono-methylation. MH_Mod is the mass of heavy mono-methylation. ZPre is the charge state of the precursor). The fragment peaks of the deduced spectrum are generated from the potential light- and heavy-spectrum as follows: both spectra are deisotoped and filtered with 10 peaks per 100 Da filter, normalized with base peak intensity. A m/z value from light-spectrum or heavy-spectrum is matched if Deltamz = (3.01883×NPeak_Mod + 4.022185×NPeak_Met) /ZPeak (NPeak_Mod ϵ [0, NMod] is the number of methyl considered in the fragment peak. NPeak_Metϵ [0, NMet] is the number of methionine considered in the fragment peak. ZPeak ϵ [1,2] is the charge of the fragment peak). If a peak in one spectrum could match multiple peaks in another, only the peak pair with minimum intensity difference is retained. The m/z of fragment without methyl is calculated from the matched pair peaks following the equation MZPeak=MZPeak_L–NPeak_Mod×ML_Mod/ZPeak or MZPeak=MZPeak_H-(NPeak_Mod×MH_Mod + 4.022185×Nmetf)/ZPeak (MZPeak_L is the m/z of pair peak from light-spectrum. MZPeak_H is the m/z of pair peak from heavy-spectrum). In the resulting deduced spectra, only peaks corresponding to paired peaks were retained and the minimum number of matched peaks was 6. And the intensity value were retained accordingly.
Database searching
The two MGF files obtained in the previous step were searched against the human protein database from Uniprot (version 20171218) using the local Mascot Demon v 2.5.1. Carbamidomethyl on Cys, Acetyl on Protein N-term and Oxidation on Met were set as variable modifications. The mass tolerance for precursor ions and fragment were set to be 5 ppm and 0.01 Da. Tryptic cleavages at Lys or Arg was selected and up to three missed cleavage sites were allowed. After the database search, the identification results were filtered with p-value of 0.05 and only rank one PSM was retained for each spectrum. Then the following criteria were applied to further filter the result: 1) the number of methionine in the peptide sequence of a PSM is equal to NMet; 2) the potential methylation site count is no less than Nmod; 3) and the light identification and heavy identification from a generated spectrum pair is identical. Then, the obtained datasets were further filtered with FDR ≤ 1% at PSM level.
Localization of methylation site
The methylation site is located based on the phosphoRS algorithm42. The methylated peptide isoforms are generated by considering the eight potential amino acid residues and three methylation types. Then the probability that the observed match between theoretical and acquired spectrum in a random event are determined. This probability is calculated by applying the cumulative binomial distribution:
In this formula n is the number of potentially existing theoretical fragment ions, k is the number of matches between theoretical and measured peaks, and p is the probability of randomly matching a single theoretical fragment ion. The probability p for a theoretical peak matching one of the experimental peaks by chance is defined by \(p=\frac{{Nd}}{w}\)Where N is the total number of extracted peaks, d is the specified fragment ion mass tolerance, and w is the full mass range of the MS/MS spectrum. Isoform probability and site probabilities are calculated using the inverse probabilities. Methylation events with localization probability of ≥ 0.75 resulted from both isotopic forms were considered to be methylation events of high confidence.
Bioinformatics analysis
Weblogo (http://weblogo.berkeley.edu/logo.cgi) were used to analyze the sequence characteristic.
Establishment of the CARNMT1-KO cells
HeLa cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Ausbian) and 1% penicillin/streptomycin. All cells were cultured at 37 °C with saturated humidity and an atmosphere of 5% CO2 and regularly tested for mycoplasma contamination. To establish the CARNMT1 knock-out cells, the pX330 plasmids containing the CARNMT1-sgRNA were transfected into the HeLa cells. After transfection for 48 h, the cells were sorted with FACS by mCherry. Cell colonies selected were sorted into 96 well plates to amplify. Western and PCR were carried out to detect the correct knock-out clones.
Evaluation of the methylation level when CARNMT1 is deleted
pcDNA3.1 plasmids containing flag tagged ZC3H8, ZC3H15, ZC3H18 or U2AF1 were transfected in CARNMT1-WT or CARNMT1-KO HeLa cells, respectively. The transfection was performed with Lipofectamine 2000 (Thermo Fisher) following the manufacturer’s protocol. The obtained cells were lysed and the resulting lysates were enriched with Anti-FLAG® M2 Magnetic Beads. The enriched proteins were digested with trypsin and analyzed with LC-MS analysis.
Recombinant protein expression and purification
Briefly, pET24a-MBP-U2AF1(1-193) and pGEX-4T-GST-U2AF2(62-138) were co-expressed in BL21(DE3) E. coli cells to improve the stability of expressed U2AF1. pET28a-6xHis-GB1-CARNMT1(53-409) plasmids were expressed in BL21(DE3) E. coli cells. The transformed bacteria were selected on LB plates containing 50 μg/ml kanamycin or Ampicillin. The isolated colonies were grown in LB medium at 37 °C until the optical density (OD600) reached 0.8. Protein expression was induced with 0.5 mM Isopropyl β-D-thiogalactoside (IPTG) by shaking overnight at 16 °C. Cells were collected by centrifugation and then lysed by sonication. Lysates were then purified with Ni Sepharose 6 Fast Flow for His-tag Protein, Glutathione Agarose for GST-tag protein and amylose resin for MBP-tag proteins. Purified proteins were concentrated by 10 kDa MWCOs. The purified proteins were evaluated by separation on SDS‒PAGE and visualized via Coomassie blue staining.
Determination of the modification structure
Synthesized Nτ-me-histidine and Nπ-me-histidine containing peptides were isotopically labeled (dimethyl labeling following reported procedure43) with light and heavy forms respectively to differentiate each other by mass when mixed together, and subjected to LC-MS analysis to evaluate the retention time of different methylation forms. Then, the synthesized Nτ-me-histidine and Nπ-me-histidine containing peptides were isotopically labeled with light and heavy forms respectively, and spiked into the digest of the enriched protein samples that were labeled with the medium forms to evaluate the in-vivo states for 5 sites in 4 proteins (ZC3H8, ZC3H15, ZC3H18 and U2AF1). The isotopically labeled samples were also subjected to Cu-IMAC separation, and were eluted with different concentration of imidazole (wash1: 1 mM; wash2: 2 mM). the resulting fractions were desalted and analyzed with LC-MS analysis.
In vitro Methylation assay
Purified CARNMT1 was evaluated to methylate its substrates (___domain peptides or proteins). The reaction samples were prepared in buffer containing 20 mM Tris-HCl (pH 7.8), 0.1 mM Zn2+, and 1 mM DTT. The final concentration of the purified protein or ___domain peptides in each sample was 20 μM. CARNMT1 were added at a final concentration of 10 μM, respectively, and S-adenosylmethionine was added at a final concentration of 1 mM. Then all of samples were incubated at 37 °C for 2 hours. The methylation of ___domain peptides was monitored with MALDI-TOF and the methylation of proteins was monitored with LC-MS analysis.
Determination of binding affinity using MST
RNAs labeled with cy5 (Supplementary Table 11) were custom purchased from Sangon Biotech. In the interaction assays, the concentration of cy5-labeled RNA was kept constant at 20 nM and the concentrations of ___domain peptides or proteins were gradient-diluted (4E + 04 nM to 19.5 nM for U2AF1 protein; 1E + 05 nM to 48.8 nM for ___domain peptides). After a short incubation, the samples were loaded into MST standard treated glass capillaries. Measurements were performed at 25 °C in buffer containing 20 mM Tris pH 7.4, 0.5 mM TCEP and 150 mM NaCl. The assays were repeated three times for each affinity measurement. Data analyses were performed using the Nanotemper Analysis and Origin software provided by the manufacturer.
RNA isolation, reverse transcription, and PCR analysis
Total RNAs were extracted with TRIzol (Invitrogen). cDNAs were synthesized from 1 μg of RNAs with oligo(dT)20VN primer using HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme). PCR was carried out using 2×Taq Master Mix (Quick Load) (Novoprotein) according to the manufacturer’s instructions. Primer sequences used in this study are listed in Supplementary Table 12.
Protein immunoprecipitations
For immunoprecipitations followed by western blot, 106 cells were harvested and suspended in the lysis buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% Triton, 1 mM DTT, 1 mM PMSF). After sonication and centrifugation, the lysate was incubated with antibody-crosslinked beads at 4 °C overnight. The beads were washed four times with the lysis buffer, and proteins were eluted for western blot analysis.
eCLIP-seq
U2AF1 eCLIP-seq experiments were performed at least in duplicate, as ENCODE guidelines (https://www.encodeproject.org), according to the published protocol32 with the following modifications: WT and CARNMT1 knock-out cells were UV-crosslinked at 400mJ/cm2. Crosslinked cell pellets were lysed in eCLIP lysis buffer and sonicated, then RNA was partially digested with RNase I (Invitrogen). Immunoprecipitation of RNA-protein complexes were performed with 10 μl anti-U2AF1 antibody (gifted from Dr Robin Reed) and Dynabeads Protein G (Invitrogen), followed by RNA linker ligation. U2AF1-RNA complexes were isolated by SDS-PAGE and transferred to nitrocellulose membranes. For U2AF1 eCLIP-seq, membrane region between 35-110 kD was excised to obtain the U2AF1 plus bound RNA. Excised membranes were treated with proteinase K and the RNA was isolated using the RNA Clean & Concentrator-5 kit (Zymo Research) according to manufacturer’s instructions. Reverse transcription and library preparation were carried out according to the published protocol32.
RNA-seq
4 μg of total RNAs were extracted with TRIzol and used for polyA+ RNA selection. Stranded cDNA libraries were generated with Universal V8 RNA-seq Library Prep Kit for Illumina Kit (Vazyme) according to the manufacturer’s protocols. The libraries were then sequenced on an Illumina Novaseq using a double-read protocol of 300 cycles at, Shanghai, GENEWIZ., INC, Suzhou, China.
Alternative splicing analysis
The RNA sequencing reads were aligned to the human genome (UCSC genome browser hg19) using STAR44 with the following parameters (parameters --chimSegmentMin 2 --outFilterMismatchNmax 3 --alignSJDBoverhangMin 6), and PSI values were calculated using Rmats45. To identify CARNMT1 KO-related AS events and U2AF1 KD-related AS events, we compared the treatment samples with the control samples and used a strict cutoff of |∆PSI | > 0.2 and FDR < 0.01. For each sample, we also used SPLICE-q46 to quantify the splicing efficiency.
eCLIP-seq analysis
eCLIP-seq reads were processed using FastUniq47 for duplicate removal and Cutadapt48 for adapter trimming. After quality control, the reads were aligned to the human genome with STAR. BAM files were converted into BED files through bedtools to extract the genomic position of the cross-linked nucleotide. They were then re-converted into single nucleotide BAM files for post-processing analysis. For each single nucleotide binding site, we normalized the binding strength using the binding reads of input samples. The binding strength around 3’ splice sites (from −20 to +5) were plotted.
Analysis of CARNMT1 using TCGA database
The gene expression level data of CARNMT1 and the clinical data were downloaded from Genomic Data Commons (https://portal.gdc.cancer.gov). For the survival analyses, we ranked the CARNMT1 expression level in all kidney cancer patients and grouped the patients in the top and bottom quartiles into “CARNMT1 high” and “CARNMT1 low” groups. The comparison of the overall survival between these two groups was performed using Cox regression in the R package “survival” and “survminer.”
Preparation of the structures for molecular simulation analysis
The structure of human U2AF1/RNA complex29 (included Zn2+) was obtained from Protein Data Bank (PDB ID:7C06(UAGGU), 7C07(AAGGU), https://www.rcsb.org/). The zinc finger structures that could be methylated, as well as the zinc finger structures that cannot be methylated was obtained from AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk)49. The complex of different zinc fingers with Zn2+ were built by overlaying the structures from AlphaFold Protein Structure Database onto the U2AF1/RNA structure. Then, the histidine which constituting C3H1 in different zinc finger structures (included U2AF1) were modified to methylated histidine to construct the methylated U2AF1/RNA/Zn2+ structure and C3H1 zinc fingers structures.
Molecular dynamics simulations
Atomistic molecular simulations were prepared on human U2AF1/RNA complex and different C3H1 zinc fingers in resting and methylation states using AMBER16 software (https://ambermd.org/), embracing Amber DNA.bsc1 force field50 for RNA and Amber ff14SB51 force field for protein. The parameters of methylated histidine were calculated using the Gaussian 09 program with the B3LYP functional under 6–311 G* basis set. The partial charges of methylated histidine were derived using the RESP charge fitted with the antechamber module in Amber 16. The interaction between Zn2+ and protein were handled according to the Zinc Protein Simulations project reported by Pang52,53. Each human U2AF1/RNA complex and different C3H1 zinc fingers was put in a hexahedral solvent box with a distance of 10 Å from the surface of the protein to the edge of the box, which was immersed with TIP3P54 water molecules. Then, each simulation system was neutralized with a number of sodium and chloride ions. Energy minimization was conducted by adding a strong restraint on U2AF1/RNA complex and different C3H1 zinc fingers, which was followed by minimizing the whole system with a few thousand steps. Subsequently, the whole system was heated to 300 K through conducting a 5 ns NVT simulation55, which was followed by a 5 ns NPT equilibration run. Lastly, the 1μs production run was performed with the time step of 2 fs (each simulation system was repeated for five times). All bonds associated with hydrogen atoms were constrained with SHAKE algorithm during the production run. Electrostatic interaction was treated by Particle Mesh Ewald56 and the cutoff value of nonbonded interactions was set to 9 Å.
Binding free energy calculation
The binding free energies between U2AF1 and RNA were calculated by Molecular Mechanics Generalized Born Surface Area (MM/GBSA) method57 base on the 500 snapshots extracted from the last 100 ns MD trajectory, which was performed in AMBER16. The GB equations58 and the binding free energies of the complex systems were solved to calculate electrostatic free energy of solvation (∆GGB). The binding energy (∆GGBTOT) can be represented as follows:
where ∆GGBTOT was obtained by summing the electrostatic energy (∆EELE), van der Waals (∆EvdW) energies, nonpolar (∆GGBSUR) and contributions polar (∆GGB). The electrostatic free energy of solvation (∆GGB) was calculated by solving the GB equation. ∆EELE and ∆EvdW were calculated according to the AMBER ff14SB.
Molecular simulation data analysis
We performed the analysis of root mean square fluctuation (RMSF), radius of gyration (Rg), root mean square deviation (RMSD), dihedral, and distance between relative atoms using either CPPTRAJ59 or TCL60 scripts according to the last 100 ns trajectories of each simulation, then further processed and plotted using Python. All structures were extracted from the simulation trajectories and processed with Pymol.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data generated in this study have been deposited in the iProX database under accession code IPX0007662001 (https://www.iprox.cn/). The mass spectrometry proteomics data have also been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository61,62 with the dataset identifier PXD047739. The data supporting the findings of this study are available from the corresponding authors upon request. Source data are provided with this paper.
References
Fontecave, M., Atta, M. & Mulliez, E. S-adenosylmethionine: nothing goes to waste. Trends Biochem. Sci. 29, 243–249 (2004).
Grillo, M. A. & Colombatto, S. S-adenosylmethionine and protein methylation. Amino acids 28, 357–362 (2005).
Chiang, P. K. et al. S-Adenosylmethionine and methylation. FASEB J.: Off. Publ. Federation Am. Societies Exp. Biol. 10, 471–480 (1996).
Biggar, K. K., Wang, Z. & Li, S. S. SnapShot: Lysine Methylation beyond Histones. Mol. cell 68, 1016–1016.e1011 (2017).
Wu, Q., Schapira, M., Arrowsmith, C. H. & Barsyte-Lovejoy, D. Protein arginine methylation: from enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 20, 509–530 (2021).
Martin, C. & Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 (2005).
Hamamoto, R., Saloura, V. & Nakamura, Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat. Rev. Cancer 15, 110–124 (2015).
Luo, M. Chemical and Biochemical Perspectives of Protein Lysine Methylation. Chem. Rev. 118, 6656–6705 (2018).
Xu, J. & Richard, S. Cellular pathways influenced by protein arginine methylation: Implications for cancer. Mol. cell 81, 4357–4368 (2021).
Hornbeck, P. V. et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic acids Res. 43, D512–520, (2015).
Zhang, L. et al. Cysteine methylation disrupts ubiquitin-chain sensing in NF-κB activation. Nature 481, 204–208 (2011).
Tessarz, P. et al. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 505, 564–568 (2013).
Wilkinson, A. W. et al. SETD3 is an actin histidine methyltransferase that prevents primary dystocia. Nature 565, 372–376 (2019).
Kwiatkowski, S. et al. SETD3 protein is the actin-specific histidine -methyltransferase. eLife 7, e37921 (2018).
Ong, S.-E., Mittler, G. & Mann, M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods 1, 119–126 (2004).
Wang, K. et al. Proteomic analysis of protein methylation in the yeast Saccharomyces cerevisiae. J. Proteom. 114, 226–233 (2015).
Drozak, J. et al. UPF0586 Protein C9orf41 Homolog Is Anserine-producing Methyltransferase. J. Biol. Chem. 290, 17190–17205 (2015).
Cao, R., Zhang, X., Liu, X., Li, Y. & Li, H. Molecular basis for histidine N1 position-specific methylation by CARNMT1. Cell Res. 28, 494–496 (2018).
Wang, K. Y. et al. Antibody-Free Approach for the Global Analysis of Protein Methylation. Anal. Chem. 88, 11319–11327 (2016).
Fu, M. & Blackshear, P. J. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat. Rev. Immunol. 17, 130–143 (2017).
Murn, J., Teplova, M., Zarnack, K., Shi, Y. & Patel, D. J. Recognition of distinct RNA motifs by the clustered CCCH zinc fingers of neuronal protein Unkempt. Nat. Struct. Mol. Biol. 23, 16–23 (2016).
Huttlin, E. L. et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440 (2015).
Huttlin, E. L. et al. Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Zhang, F. et al. Global analysis of protein arginine methylation. Cell Rep. Methods 1, 100016 (2021).
Dai, L. et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 10, 365–370 (2014).
Zhao, Y. M. et al. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 (2011).
Yoshida, H. et al. A novel 3’ splice site recognition by the two zinc fingers in the U2AF small subunit. Genes Dev. 29, 1649–1660 (2015).
Yoshida, H. et al. Elucidation of the aberrant 3’ splice site selection by cancer-associated mutations on the U2AF1. Nat. Commun. 11, 4744 (2020).
Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Shao, C. et al. Mechanisms for U2AF to define 3’ splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 21, 997–1005 (2014).
Van Nostrand, E. L. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016).
Jerabek-Willemsen, M. et al. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 1077, 101–113 (2014).
Yang, F., Shen, Y., Camp, D. G. 2nd & Smith, R. D. High-pH reversed-phase chromatography with fraction concatenation for 2D proteomic analysis. Expert Rev. Proteom. 9, 129–134 (2012).
Shimazu, T. et al. Histidine N1-position-specific methyltransferase CARNMT1 targets C3H zinc finger proteins and modulates RNA metabolism. Genes Dev. https://doi.org/10.1101/gad.350755.123 (2023).
Biancon, G. et al. Precision analysis of mutant U2AF1 activity reveals deployment of stress granules in myeloid malignancies. Mol. Cell 82, 1107–1122.e1107 (2022).
Shirai, C. L. et al. Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat. Commun. 8, 14060 (2017).
Esfahani, M. S. et al. Functional significance of U2AF1 S34F mutations in lung adenocarcinomas. Nat. Commun. 10, 5712 (2019).
Metz, P. J. et al. Symmetric Arginine Dimethylation Is Selectively Required for mRNA Splicing and the Initiation of Type I and Type III Interferon Signaling. Cell Rep. 30, 1935–1950.e1938 (2020).
Cancer Genome Atlas Research, N. et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Teleman, J., Chawade, A., Sandin, M., Levander, F. & Malmstrom, J. Dinosaur: A Refined Open-Source Peptide MS Feature Detector. J. proteome Res. 15, 2143–2151 (2016).
Taus, T. et al. Universal and confident phosphorylation site localization using phosphoRS. J. proteome Res. 10, 5354–5362 (2011).
Boersema, P. J., Raijmakers, R., Lemeer, S., Mohammed, S. & Heck, A. J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Shen, S. H. et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 111, E5593–E5601 (2014).
Costa, V. R. M., Pfeuffer, J., Louloupi, A., Orom, U. A. V. & Piro, R. M. SPLICE-q: a Python tool for genome-wide quantification of splicing efficiency. Bmc Bioinforma. 22, 368 (2021).
Xu, H. B. et al. FastUniq: A Fast Duplicates Removal Tool for Paired Short Reads. Plos One 7, e52249 (2012).
Kechin, A., Boyarskikh, U., Kel, A. & Filipenko, M. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput Biol. 24, 1138–1143 (2017).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583 (2021).
Ivani, I. et al. Parmbsc1: a refined force field for DNA simulations. Nat. Methods 13, 55 (2016).
Maier, J. A. et al. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput 11, 3696–3713 (2015).
Pang, Y. P. Successful molecular dynamics simulation of two zinc complexes bridged by a hydroxide in phosphotriesterase using the cationic dummy atom method. Proteins 45, 183–189 (2001).
Pang, Y. P., Xu, K., El Yazal, J. & Prendergast, F. G. Successful molecular dynamics simulation of the zinc-bound farnesyltransferase using the cationic dummy atom approach (vol 9, pg 1857, 2000). Protein Sci. 9, 2583–2583 (2000).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 79, 926–935 (1983).
Berendsen, H. J. C., Postma, J. P. M., Vangunsteren, W. F., Dinola, A. & Haak, J. R. Molecular-Dynamics with Coupling to an External Bath. J. Chem. Phys. 81, 3684–3690 (1984).
Darden, T., York, D. & Pedersen, L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 98, 10089–10092 (1993).
Xu, L., Sun, H. Y., Li, Y. Y., Wang, J. M. & Hou, T. J. Assessing the Performance of MM/PBSA and MM/GBSA Methods. 3. The Impact of Force Fields and Ligand Charge Models. J. Phys. Chem. B 117, 8408–8421 (2013).
Onufriev, A., Bashford, D. & Case, D. A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins-Struct. Funct. Bioinforma. 55, 383–394 (2004).
Roe, D. R. & Cheatham, T. E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput 9, 3084–3095 (2013).
Humphrey, W., Dalke, A. & Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph Model 14, 33–38 (1996).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic acids Res. 47, D1211–D1217 (2019).
Chen, T. et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic acids Res. 50, D1522–D1527 (2022).
Acknowledgements
This work was supported, in part, by funds from the China State Key Basic Research Program Grants (2021YFA13026012 [M.Y.], 2019YFA0709400 [G.L.], 2022YFA1303300 [H.C.]), the Strategic Priority Research Program of Chinese Academy of Sciences (XDB37040401 [G.L.], XDB0570100 [H.C.]), the National Natural Science Foundation of China (21804131 [K.W.], 92153302 [M.Y.], 21933010 [G.L.], 31925008 [H.C.], 32230022 [H.C.], 32430023 [H.C.]), the innovation program (DICP I202226 [K.W.]) of science and research from the DICP, CAS.
Author information
Authors and Affiliations
Contributions
M.Y., K.W., and H.C. conceived and designed the project. K.W. carried out the methylation identification experiments and MS analysis under the supervision of M.Y. K.W. and J.M. analyzed the MS data. K.W., L.Z. and Z.L. performed the cellular and biochemical experiments with help from L.X., J-S.W. and K.L. Y.L. and G.L. conducted the molecular simulation analysis. S.Z. and Z.W. conducted the data analysis of RNA-seq and eCLIP. K.W., Y. M. and J-Y.W. conducted the in-vitro RNA binding experiments. H. L. participated in the confirmation of the responsible enzyme for C3H1 zinc fingers. K.W., M.Y., and H.C. wrote the manuscript with input from L.Z., Y.L., S.Z. and J.M.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Chao Xu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, K., Zhang, L., Zhang, S. et al. Metabolic labeling based methylome profiling enables functional dissection of histidine methylation in C3H1 zinc fingers. Nat Commun 15, 7459 (2024). https://doi.org/10.1038/s41467-024-51979-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51979-2