Abstract
The dog, Canis lupus familiaris, is an important model for studying human diseases. Unlike many model organisms, the dog genome has a comparatively poor functional annotation, which hampers gene discovery for development, morphology, disease, and behavior. To fill this gap, we established a comprehensive tissue biobank for both the dog and wolf samples. The biobank consists of 5485 samples representing 132 tissues from 13 dogs, 12 dog embryos, and 24 wolves. In a subset of 100 tissues from nine dogs and 12 embryos, we characterized gene expression activity for each promoter, including alternative and novel, i.e., previously not annotated, promoter regions, using the 5’ targeting RNA sequencing technology STRT2-seq. We identified over 100,000 promoter region candidates in the recent canine genome assembly, CanFam4, including over 45,000 highly reproducible sites with gene expression and respective tissue enrichment levels. We provide a promoter and gene expression atlas with interactive, open data resources, including a data coordination center and genome browser track hubs. We demonstrated the applicability of Dog Genome Annotation (DoGA) data and resources using multiple examples spanning canine embryonic development, morphology and behavior, and diseases across species.
Similar content being viewed by others
Introduction
Since the annotation of its genome a decade ago, dogs have proven to be an excellent model for studying inherited diseases. Many spontaneous, simple, and complex phenotypes occur in dogs, providing physiologically relevant models for corresponding human conditions. A unique population history and breed structure have resulted in a genetic architecture that facilitates the exploration of the genetic basis of simple and complex disorders with smaller study cohorts and fewer markers than in humans. Numerous examples exist across disease groups, including epilepsy1, kidney disease2, developmental conditions3, skin diseases4, eye disorders5, and cancer6,7. Fascinating discoveries have also been made across morphological traits and appearance8,9,10,11, and a growing number of loci and candidate genes have been suggested for behavior and anxiety, overlapping genomic loci in human psychiatric disorders12,13,14.
A widely used reference genome for the dog, CanFam3.1, was generated by sequencing the DNA of the Boxer Tasha in 2005, with improvements in 201415,16. This was a significant step forward in canine genetic research, enabling the development of efficient gene mapping tools. However, the CanFam3.1 reference still contained over 20,000 gaps, missing promoters, and other regulatory elements. Recent long-read sequencing efforts have addressed these challenges and led to massive continuity improvements, removing most gaps and producing high-quality reference genomes in different breeds and dingo17,18,19,20,21.
Despite the improved reference genomes, dog genome annotation remains far from complete. Regulatory elements such as transcription start sites (TSSs), promoters, and enhancers, which are crucial for gene expression and often affected by complex traits, remain uncharacterized22. To address this challenge, we formed the Dog Genome Annotation (DoGA) Consortium to generate functional annotations by establishing a large tissue biobank of dogs and wolves, identifying and annotating functional regions using 5’ technologies, setting up a gene expression atlas across 100 tissues and generating community resources to browse and utilize the data. This extraordinary effort should further facilitate gene discovery in regulatory regions and improve our understanding of gene regulation in dogs, wolves, and humans.
Results
A large canine tissue biobank
We initiated the DoGA project for promoter and enhancer annotation employing targeted RNA sequencing technologies such as STRT2-seq and CAGE-seq (separate, ongoing manuscript) (Fig. 1).
To establish a high-quality tissue biobank, we collected samples from 49 animals, including 13 dogs, 12 dog embryos, and 24 wolves (including 11 adults and 13 pups) (Fig. 1, Supplementary Data 1). We performed a thorough clinical examination of the recruited animals, followed by necropsy and histological examination of the dissected tissues to ensure that only tissue samples without histological changes were selected for subsequent analyses (three dogs were excluded; see Methods and Supplementary Fig. 1,2). We targeted 132 tissues from 13 organ systems, i.e., of related organs, see Fig. 2. Multiple samples were collected from several organs to separate the different anatomical parts; for example, 22 brain regions were sampled. Overall, 5926 samples (1850 unique samples, i.e., excluding replicates) were stored in our biobank and will be available to the scientific community. The biobank will grow with new canine recruitments.
Bold tissue names have been sequenced with STRT2-seq. In brackets, we indicate the number of enriched robust promoters, genes, and their tissue-enriched orthologs in the Human Protein Atlas for the specific tissue and organ system. Dashes indicate no orthologue tissue was included in the Human Protein Atlas.
Tissue-level 5’-expression atlas
To simultaneously capture the active promoter regions and quantify gene expression levels, we utilized the low-input STRT2 sequencing approach. A QC report for STRT2 sequencing with MultiQC report with statistics from FastQC and HiSAT2 is included in Supplementary Data 2. Compared to most other 5’-prime sequencing methods, such as CAGE-seq23, STRT2 enabled us to discover promoters even in tissues with a low abundance of RNA, such as the retina and the adenohypophysis. This allowed us to generate data from a large set of individual tissues in our biobank, from nine dogs and 12 embryos. We studied 361 samples from 100 tissues and 13 organ systems for a broad representation of dog physiology, emphasizing several regions of the central nervous system (CNS) (Fig. 2). To obtain reliable transcriptomic data, we sampled at least three healthy animals for nearly all target tissues in biological replicates, including reproductive tissues from both sexes (Supplementary Data 1).
Identification of 100,000 promoters in 100 tissues
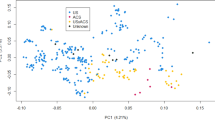
Promoter regions were annotated using a method similar to that used by the FANTOM5 consortium24. We identified 106,681 promoters in the 100 tissue samples, called the comprehensive promoter set (Fig. 3a). Promoters expressed in at least three biological replicates were put into a robust set of 46,644 regions. We further classified these promoters based on their overlap with annotated genes in CanFam4 (NCBI v106, Table 1). Among the robust promoters, we report 15,654 promoters, which are in ±500 bp proximity to the TSSs of CanFam4 genes, including 16,596 alternative promoter sites and 14,394 novel promoter candidates. The promoter sites in our samples show similar expression profiles according to their annotated biological origin based on their organ systems and tissues (Fig. 3b). Six major groups are forming in the plot, which can be described by their biological function. For example, all samples from different brain regions group together with retinal samples based on the expression of the promoters. Another group comprises all muscular tissue samples, except cardiac muscle, spanning different organ systems. One of the six groups contains a mixture of different tissues with subgroups with specific biological meaning, e.g., all non-retinal eye samples. We studied the impacts of biological factors, such as sex, age, and breed, based on visual inspection of the UMAP plots and did not observe any discernible effects (Supplementary Fig. 3). The overall grouping holds even when looking at expression profiles of primary, alternative, or novel promoters (Supplementary Fig. 4).
a Overview of our different promoter classes and their sizes. b The UMAP plot of the STRT2-seq tissue samples based on the expression of the robust promoter set. Each dot represents one sample and was color-coded based on their organ system of origin in the metadata. Overlapping causes the darkening of dots. The neuronal tissue cluster consists of CNS samples as well as retinal tissue. c The number of tissue-enriched robust promoters in each tissue (right) and the number of organ-system-enriched robust promoters in each organ system (left).
Validation of identified promoters
To further validate these promoters, we evaluated the level of support for each promoter class using public epigenomic data from a subset of tissues. As expected, the promoters were coinciding to a large degree with open chromatin regions (45–55% overlap) and with the active promoter marks H3K4me3 (25–35%) and H3K27ac (47–55%)12 when remapped to CanFam4.1 (Table 1). When looking at the different promoter types, we see the highest overlap with epigenetic marks for primary promoters, then alternative, and the lowest for novel promoters.
To validate some of our new promoter candidates, we investigated the transcripts of five robustly expressed candidate genes, ATP5ME, MTIF3, TNNI3, TBC1D22B and GPT, with putative new first exons or TSSs based on gene models annotated in CanFam3.1, by RT-PCR and Sanger sequencing with several different primers. The sequences were merged to create the consensus sequence for each cDNA. According to the UCSC’s BLAT search on CanFam4.1, all except one (TNNI3) of the new CanFam3.1 TSSs were present in CanFam4.1, indicating the improved canine reference and validating our STRT2-based findings. The new TNNI3 transcript TSS is located upstream of exon 3 (Supplementary Data 3).
Tissue enrichment of genes and their promoters
Promoters and genes are variably expressed in different tissue types24. To capture this, we examined the expression levels of robust promoters and genes (mapped to CanFam4) using an approach similar to what was employed in the Human Protein Atlas Project25. We detected ubiquitous and tissue-enriched expression of the promoters and genes. We found 9367 genes and 12,505 robust promoters enriched in one or more tissues, with the testis having the most enriched promoter regions (6626) and genes (6326), as was the case in humans (Fig. 3c). By comparing organ systems, we identified 9108 robust promoters and 4770 genes that were enriched in a specific organ system. The embryonic system had the most enriched genes (1825); in contrast, the central nervous system had the highest number of enriched robust promoters (1779) (Fig. 3c). Of the 12,505 tissue-enriched promoters, 4675 were identified as alternative and 3254 as novel, i.e., not annotated promoter regions. We developed an online expression atlas to explore gene and promoter expressions (https://expression-atlas.doggenomeannotation.org/dogaatlas/). This enables visualization of expression profiles across tissues based on genomic ___location, gene symbols, and tissue names and exports a list of genes of interest. The full list of tissue-enriched genes and promoters is provided in Supplementary Data 4 and 5. The expression atlas also includes the tissue-enriched genes identified by EpiC-dog and allows us to see their expression patterns in the context of our data.
Biological Significance of the DoGA resource
The following use cases indicate some possible analyses with the DoGA resource to improve biological insights.
Example use-case: expression profiles of OMIA genes
The Online Mendelian Inheritance in Animals (OMIA) database collects diseases, traits, and associated genes26. One use case for the DoGA resource is that it can provide the tissue context and prioritization for disease-associated genes. We extracted 292 monogenic canine disease records from OMIA, lifted them over from CanFam3.1 to CanFam4, and cross-referenced them with our expression profiles. We observed a clear tissue enrichment for specific disease-related genes, e.g., SIX6, which is related to an inherited eye malformation and is primarily expressed in the retina and other eye-related tissues. Other genes are utilized in several tissues. For example, the canine microphthalmia-associated27 gene RBP4 shows, besides a high expression in the retina, similar levels in the common bile duct and even higher in the liver (Fig. 4a). Not only is disease-related gene expression variable across tissues, but alternative promoters of these genes can also be expressed differently. For example, out of 11 alternative promoters for RBP4, we found 8 with an enriched expression in the liver and 3 with high expression over several tissues, including the retina (Supplementary Fig. 5).
a log-normalized expression of genes associated with eye-development-related diseases across all tissues. b Alternative promoter usage for RGS3 in cardiac muscle tissues. c Expression profiles of genes enriched during embryonic development compared to adult samples. We highlighted one gene enriched in the early stage (AIPL1) and one later stage (CBX2). d The expression of robust promoters overlapping with SNPs is associated with “Sorry when wrong” behavior.
Example use-case: conservation of tissue-enriched gene expression between dogs and humans
We compared the expression of genes and especially their tissue enrichment between dogs and humans to strengthen the suitability of dogs as models for human health and physiology. Of the 9367 tissue-enriched genes we found in dogs (CanFam4), we identified 1001 enriched 1:1 gene orthologs in humans based on data from the Human Protein Atlas25. The majority (726 genes, 73%) of these orthologs were enriched in the same tissue in both species, with testis being the most common, with 473 enriched genes.
As a case study for the sub-tissue resolution of our promoter data set, we analyzed the samples from atria and ventricles. We compared the expression patterns to a study in humans28. As seen in humans, we observed a similar high expression of NPPA in the atria and MYL2 in the ventricles. For Regulator of G Protein Signaling 3 (RGS3) gene, we observed high expression in the right ventricle for promoter 1 (RGS3_p1) and the right heart atrium for promoter 2 (RGS3_p2) (Fig. 4b). Intriguingly, the same pattern is observed in humans28.
Example use-case: overlap of promoters with lineage- and behavior-specific SNPs
Two recent studies analyzed SNPs related to different dog breed lineages and their genetic background of behavioral traits12,29. To help explain more of these variants, we cross-referenced our promoter candidates with these variants on CanFam4. Looking at the 6327 lineage-specific SNP clusters and the robust promoter set, we found 1213 primary and 1256 alternative promoters coinciding with polymorphisms, meaning a SNP cluster falls inside the region of a promoter candidate. Additionally, 1053 novel promoters cover SNPs identified in this study, allowing first steps toward potential causal explanations of these variants. Due to the larger span of certain SNP clusters, one cluster can overlap with multiple promoter regions. In total, 291 of the 6327 lineage-specific SNP clusters (7%) are covered by a promoter region. A closer look at the overlap for each lineage-specific subset is given in the expression atlas (https://expression-atlas.doggenomeannotation.org/dogaatlas/). A majority of SNPs overlapping with promoters belong to the retriever breed lineage. The same analysis for the 1079 behavior-related SNPs overlaps 240 alternative and 257 primary promoter regions in the robust set. Additional 276 novel promoter regions coincide with SNP clusters (Table 2). In total, 284 of the 1079 behavior-related SNP clusters (26%) are covered by a promoter region. The SNP group with the highest number of hits is “Sorry when wrong”-behavior. The majority of the hit promoters in this category are alternative or novel, though the expression doesn’t show any tissue-level enrichment (Fig. 4d).
Example use-case: Gene and promoter usage during dog embryonic development
Developing a fertilized oocyte into a newborn canine puppy requires ~63 days. The whole embryos used in this study were determined to be either 20–25 or 30 gestational days old. Various important developmental events take place at these time points. We found distinct quantitative and qualitative differences in the promoter usage at different developmental stages (Fig. 4c). The number of enriched genes increased from 96 to 454 in the transition from the 20–25 to the 30-day stage of embryonic development, reflecting the active formation of the body organs at 30 days. We found that known promoter activation coincides with the expected onset of the organ and tissue development, such as a known retinal development gene, AIPL1, at 20–25 days30,31. We see the strongest expression of this gene in the earlier embryos compared to older embryos or adult samples (Fig. 4c). Similarly, expression of CBX2, which is involved in gonad development and sex differentiation32, peaks at 30 days and decreases again in adults, together with other genes involved in sex differentiation (Fig. 4c).
Resources to access and utilize the DoGA data
A key goal of the DoGA consortium is to enable easy access to resources and annotations (Table 3). To this end, we established a publicly available data coordination center (DCC) at https://dcc.doggenomeannotation.org/doga (Fig. 1) to allow the exploration and retrieval of detailed information on animal samples, sequencing libraries, raw and processed data, and analysis results for CanFam3.1 and CanFam4. We developed an online expression atlas to explore gene and promoter expression (https://expression-atlas.doggenomeannotation.org/dogaatlas/). Additionally, we provide genome browser views for our data, functional annotation, and species-wide comparison using the Zenbu genome browser33 and a UCSC Genome Browser track hub34. We further provide data analysis scripts as R-Markdown documents in our DoGA Gitlab repository (https://gitlab.com/doggenomeannotation/strt) to facilitate the replication of our analysis with different reference genomes. In addition to the STRT2-seq data, we also generated whole genome sequencing data from six dogs and four wolves to allow the evaluation of allele-specific analyses of gene expression in future projects. Sequence data has been deposited in SRA (BioProject PRJNA907518), and VCF files are available in the DoGA DCC.
Discussion
This landmark study, with a wide array of high-quality resources and tools, will enable scientific discovery in dogs at an unprecedented level, with implications for the genomics community in the coming years. This biobank, promoterome, and expression atlas will facilitate research on evolutionary adaptations and gene discovery across traits in dogs, wolves, humans, and other mammals. For example, including a wide range of tissues from the CNS of both dogs and wolves will provide a unique opportunity to study the molecular basis of the behavioral evolution of domestication in mammals in the future.
Our extensive DoGA biobank combined with a low-input 5’-transcript sequencing technology STRT2 produced a major complementary data set for the functional annotation of the canine genome. Concerning previous efforts in dogs, this study is the first comprehensive functional annotation for CanFam4, improving the quality of previous annotation in CanFam3.1 by providing the transcription-derived base pair-resolution promoterome compared to the previously described RNA-seq transcriptome21 and histone mark-based promoter regions35,36. This study includes 100 distinct tissue types and is more extensive than previously published data sets in BarkBase (27 tissue types) and EpiC Dog (11 tissue types). For example, our 22 subregions in the CNS versus two brain regions in EpiC Dog allow for more in-depth studies of neuronal gene expression and regulation. Similarly, our whole embryo samples (20–30 days old) complement the older embryonic tissues (33–44 days old) published in BarkBase, providing insights into promoter usage at the onset of organ development, which starts at around 30 days. Compared to RNA-seq or ChIP-seq-based sequencing technologies, STRT2-seq gives base pair-resolution of primary and alternative TSSs and the expression level of the corresponding genes23. Additionally, the low input requirements of this technique allowed us to study a wide array of small organ regions and tissue types. However, the goals and technologies of DoGA, EpiC-dog, and BarkBase are different and provide complementary insights into the functional annotation of the dog genome. For example, our data can support the tissue-enriched genes identified in EpiC-dog and show their expression in the broader tissue context.
We discovered 107,000 promoters in the comprehensive set, including 41,000 novel promoters, i.e., promoters not in close proximity to any hitherto annotated genes. The large number of novel promoters is not surprising, based on our study’s wider range of tissues and the 5’ sequencing approach, which was also the case in previous studies in humans24. Therefore, our DoGA resource and functional annotation can serve diverse research questions, such as addressing the role of conserved regulatory elements, alternative promoter usage in different tissues and developmental stages, allele-specific transcription combining genomic and transcriptomic data, and facilitating the prioritization of candidate causal variants across traits. Accordingly, the DoGA resources have been recently utilized to study expression patterns or regulatory variants in dog color patterns37, eye disease38, dilated cardiomyopathy39 and additional efforts are ongoing across traits.
We analyzed the quality of our identified promoter candidates in several ways. First, they show a high overlap (35–55%) with promoter-associated histone marks in selected tissues available from previously published data sets. Second, we validated several promoter candidates using PCR. Additionally, the grouping of the expression of our samples according to their biological function in the UMAP plot (Fig. 3b) and the development-specific expression of promoters during different time points in embryonic samples demonstrate the high quality of our tissue sampling and data generation.
This study highlights additional use-case examples for our resources. We showed tissue-specific expression profiles for hundreds of OMIA records, demonstrating the utility of the diverse tissue collection to identify relevant tissues and genes affected by disease- and trait-related genomic variations. The variation in expression of alternative promoters in different tissues, as seen in RBP4, could help explain the effects on tissue-specific disease expression. We revealed the conservation of tissue-enriched expression between dogs and humans in 1001 different genes. As shown with the example of cardiac tissues, our data also allows the analysis of the conservation of alternative promoter usage between humans and dogs. Additionally, we overlapped promoters with recently discovered lineage- and behavior-specific SNPs12,29 and indicated that over 1000 hitherto unknown promoter regions overlap with a SNP cluster, which will help to prioritize these loci for functional studies to prove causality. This is important not only in the genomic context of the loci, but our data also allows us to combine it with their tissue-level expression patterns. For example, we provide the ability to study the expression of promoters overlapping behavioral SNPs across different brain regions.
We also demonstrated striking developmental stage-timed gene and promoter usage changes in dog embryos from 20–25 and 30 days. The three germ layers are formed in the implanted embryos, and organogenesis starts with a detectable heartbeat and limb buds at 20–25 days30. By 30 days, the canine embryo continues to grow rapidly with the major organs and systems of the body formed, including the ossification of the bones and sex differentiation. We found a significant increase in the number of enriched genes and promoter regions in the comparison of 20–25 day embryos and 30 day-old embryos, reflecting the extensive developmental changes.
Although we provide a comprehensive resource to the scientific community, we recognize limitations. Our gene expression atlas and the list of promoters are incomplete and may lead to inconclusive results. We aimed to sample three healthy biological replicates, but our canine recruitment relied on privately owned dogs with health issues and behavioral abnormalities, which we mitigated through clinical pathological examinations and exclusion criteria. We also lack single-cell resolution, which limits our ability to distinguish different cell types and possible cell type-specific promoters in a single tissue. Additional tissues from different developmental and life stages would also reveal more promoters, further improve the functional annotation, and reveal a different set of genes and promoters enriched in just one tissue or time point. Due to difficulties extracting enough undegraded RNA, we miss promoters and expression profiles from certain tissues, including the intestine. Deeper sequencing of the STRT2-seq libraries could also have revealed additional lowly expressed promoters due to the enrichment of truncated transcripts that may interfere with identifying the biologically relevant TSSs. Our categorization of promoters into primary, alternative, and novel relied on the distance to annotated genes and may, therefore, change with improved gene models or the choice of thresholds. The difficulty in grouping nearby novel promoters into larger genomic regions hindered us from classifying them as primary and alternative. This remains one of the future tasks. Our fine-grained sampling of CNS tissues and their close relationship with each other might lead to missing enriched genes and promoters, e.g., the four different sections of the cortex compared to other tissues such as testis. Our approach to gene and promoter conservation was based on gene symbols, but novel orthologue identification methods, such as TOGA40, can be used for a more precise analysis.
In summary, we provide data and methods to annotate a growing number of reference and de-novo canine genomes. This is important since highly accurate promoter prediction and identification of alternative promoter sites will facilitate the interpretation of the causality of functional genomic variants. Similar to the FANTOM5 promoter atlas for humans, our comprehensive canine transcriptomic data will powerfully complement these efforts by providing genome-wide promoter locations with base pair resolution and usage across multiple tissues. Our data improves the accuracy of dog genome annotation and, therefore, the usability of dogs as a large spontaneous model for human health, mammalian evolution, and canine veterinary research. Utilizing these resources, the DoGA consortium will continue efforts to reveal more regulatory mechanisms in the canine genome, such as enhancers, and to compare gene regulation differences in the brains of dogs and wolves.
Methods
DoGA biobank
The DoGA Biobank contains nearly 6000 samples from up to 132 different RNA-later or snap-frozen tissue types collected from 49 animals, including 25 dogs and 24 wolves of different ages, and diverse demographic, clinical and pathological metadata (Fig. 1). Details for all animals, tissues and metadata can be found in the DoGA Data Coordination Center (DCC, https://dcc.doggenomeannotation.org/doga).
Animals
The dog samples for this project were obtained from pet dogs euthanized by the owner’s decision (information about each dog reported in DoGA DCC). A thorough clinical examination was performed on all dogs. If the neurological disease was suspected, a neurological examination was conducted using an ECVN diplomate (including assessment of mental status, posture, gait, postural reactions, spinal reflexes, and cranial nerves). Blood samples for DNA isolation and complete blood count (CBC), plasma for metabolomics analysis, and serum for biochemical analyses were obtained before euthanizing the animals. The dogs were euthanized by first sedating the animals with intramuscular dexmedetomidine and butorphanol, followed by intravenous administration of propofol and an overdose of pentobarbital. The Ethical Committee of the Faculty of Veterinary Sciences evaluated and accepted the animal proceedings under ethical permissions ESAVI/7482/04.10.07/2015, ESAVI/343/04.10.07/2016 and ESAVI/25696/2020.
Postmortem examination, tissue collection, and histological preparation
The post-mortem tissue sampling was organized into six workstations, with one sampler (pathologist) and two assistants simultaneously sampling many organs. Gross procedures included initial decapitation and exsanguination through the opening of the femoralis and jugularis muscles. Supplementary Fig. 2 shows an overview of the tissue collection stations. The head and eyes were further processed at stations 1 and 2. The right front limb was detached for further preparation at station 6. After a ventral midline incision through the linea alba, the liver and gastrointestinal tract were removed to enable sampling at station 4, separate from the animal carcass, where sampling at stations 3 and 5 took place. Tissue pieces of appr. a 1 x 1 x 1 cm3 sample were excised from the organ/sampling site according to a predefined schema for every station and further processed on cooled preparation boards into 1 x 1 x 1 mm3 samples for RNA isolation (distributed in four tubes containing RNAlater or snap-frozen in liquid nitrogen). Macroscopic changes in the body and organs were recorded during organ preparation and sampling. Tissue slices, appr. ½ × 1 × 2 cm3 in size were obtained from the ___location next to the RNA sample site and immersion-fixed in 10% buffered formalin for a minimum of 24 h. For small, paired organs (thyroid and parathyroid), the contralateral side was sampled for histology. The hypophysis was dissected into neuro- and adenohypophysis and submitted for RNA extraction or histology. Autopsy and sampling were performed within 90 min after death.
The formalin-fixed tissues were sectioned, paraffin-embedded, cut into 4 µm thick sections, mounted onto glass slides, deparaffinized with xylene, rehydrated in a decreasing alcohol series, and stained with hematoxylin and eosin (HE). Calcified and osseous tissues were cut using a diamond bandsaw (EXAKT 30/917). Fixed tissue sections were decalcified in EDTA, thoroughly rinsed in tap water for 24 h, and further processed as described above for staining. The slides of all sampled tissues were evaluated in HE-staining by a board-certified veterinary pathologist (PS) and defined using terminology standardized by the European College of Veterinary Pathologists (https://www.ecvpath.org/). Special stains (rubeinic acid for copper Masson’s trichrome for collagen/fibrosis) were used when necessary to verify the morphological diagnosis. Only tissues with no histological changes were used for RNA extraction. Regarding the liver samples, mild vacuolar degeneration and/or scattered lymphocytes within the parenchyma were accepted for further processing if the biochemical liver parameters (ALAT, ASAT, ALP) were within the normal range according to the blood sample at euthanasia.
In collaboration with the Ranua Wildlife Park (Finland) and Ähtäri Zoo (Finland), we collected postmortem tissue samples from 9 adult wolves, 13 wolf cubs at 2–9 days of age that were euthanized for management purposes (see Fig. 1). The dissection room was prepared at on-site facilities, with two pathologists and three assistants performing the tissue dissection. Brain sampling was equal to that used for dogs; extra neural tissues were limited to soft tissues. The brains of the cubs were cut in half, snap-frozen in liquid nitrogen, and stored at −80 °C until dissection. Before dissection, the brains were thawed slightly. A total of about 40 tissues per adult animal and seven from the cubs were sampled and stored in RNAlater, dry ice, or liquid nitrogen. The wolf tissue samples were processed and histologically evaluated, similar to dog samples. All the detailed information for each individual can be found on the DoGA DCC.
We also obtained brain tissue and RNA samples from two Russian adult wolves (19 tissue samples per animal). The use of Russian wolf brain samples was reviewed and approved by the local ethics committee of Kazan Federal University (Protocol No. May 9 5, 2018). Brain samples were obtained in the frame of an annual “Tissue-sharing for research needs Initiative” (season 2017–2018) by the State Committee for Biological Resources of the Republic of Tatarstan in collaboration with the Hunters Association of Tatarstan. In addition, we collected skin explants for culturing fibroblasts from 11 dogs and 2 Finnish wolves from our cohort. We also received RNA samples of 12 whole dog embryos.
RNA isolation
The tissue samples in RNAlater were placed at +4 °C for 24 h, the RNAlater was removed, and the tissue samples were stored at −80 °C. Snap-frozen tissue (N2) sections were stored at −80 °C.
RNA isolation was performed using QIAZOL (Qiagen, Germantown, MD, USA), the Rneasy Mini kit (Qiagen, Germantown, MD, USA), or a combination of both methods, according to the manufacturer’s instructions. Modifications to the homogenization protocol or incubation times were introduced when necessary. Harder tissues such as muscle and heart required more homogenization cycles (using two cycles, 6000 rpm, 15 sec/cycle), and for some tissues, three cycles were done or for a longer time (~30 s each cycle). Detailed information for every tissue is available in the DoGA database. RNA concentration and purity were measured using DeNovix D-11 (DeNovix Inc., Wilmington, USA), and the concentration was determined using a Qubit™ RNA BR Assay Kit (Invitrogen) and read on a Qubit 2.0 Fluorometer (Invitrogen). RNA integrity was determined using Bioanalyzer2100 or TapeStation4200 (Agilent, Santa Clara, CA, USA), and RNA samples with RIN values >7.0 were accepted for further analysis.
STRT2 library preparation method
We have modified Single-Cell Tagged Reverse Transcription (STRT)41,42 to suit the bulk RNA template. The starting material was 40 ng RNA per sample, and each library consisted of 48 samples. Our modifications included longer UMIs (Unique Molecular Identifiers, 8 bp instead of 6 bp) for molecular counting and Globin-Lock® to deplete globin mRNAs that are abundant in blood and blood-rich tissues dominating all reads.
Briefly, template RNA was mixed with ERCC Spike-in RNA, which is added equally to each sample for normalization, and with globin-LNA primers, which hybridize to the sequence adjacent to the poly-A+ tail of globin mRNA, thus blocking its transcription. First-strand cDNA synthesis was done using an oligo(dT) primer with the addition of 3-6 cytosines. Template switching oligo with UMIs promotes template switching and introduces UMIs into the cDNA. The cDNA was amplified (10 cycles), and the well-specific barcodes were introduced at the 5′end. The samples were then pooled and ligated to an adapter cassette. The 5′end was further amplified (10–12 cycles) for sequencing.
The quality of the library preparation was analyzed with TapeStation, and the concentration was determined with the KAPA Library Quantification Kit. The libraries were sequenced with NextSeq 500 using a High Output v2 kit, 75 cycles. The protocol details are described in Ezer et al. 43. Previous studies have shown the unbiased performance of STRT2 in GC-rich regions44,45,46,47,48,49,50.
STRT2 data processing
STRT2 data were processed as described previously51,52. Briefly, Illumina base call (BCL) files were demultiplexed using Picard (version 2.10.10; http://broadinstitute.github.io/picard/), ExtractIlluminaBarcodes, and IlluminaBasecallsToSam to generate the unaligned BAM files. The BAM files were then converted to FASTQ files with Picard SamToFastq and aligned to the dog reference genome CanFam3.1 (GCA_000002285.2) and UU_Cfam_GSD_1.0/CanFam4 (GCA_011100685.1) and ERCC spike-ins (SRM 2374) by HISAT2 (version 2.1.0)53. The aligned BAM files were then merged with the original unaligned BAM files to generate UMI-annotated BAM files using Picard MergeBamAlignment. The BAM files corresponding to each sample from different lanes were then merged using Picard MergeSamFiles. Potential PCR duplicates were marked with Picard ‘Markduplicates’. See Supplementary Data 2 for a detailed QC report and Supplementary Fig. 6 for a correlation analysis of different samples.
STRT2 tag clusters, quantification and promoter classification
The 5’-end of the mapped STRT2 reads was grouped into clusters with Paraclu54, as used by FANTOM524. For the promoter, clusters overlapping the last exon ±500 bp of annotated genes were excluded using the NCBI Canis lupus familiaris Annotation Release 106. We calculated gene-level expression by summing the expression of all clusters overlapping the annotated gene bodies. The counts were normalized to ERCC spike-in counts using logNormCounts with transform = “none” in the SingleCellExperiment package (v 1.16.0)55. The resulting clusters were then filtered to contain a normalized count higher than 50 over all samples. Clusters with a normalized count of > 20 in at least one sample were categorized as comprehensive promoters. Clusters with counts higher than 30 in at least three samples were categorized as robust promoters. Promoters in both sets were then categorized as novel if they were not in a 500 bp window around the annotated genes. In cases with multiple promoters closer than 500 bp to annotated regions, the cluster with the highest expression was categorized as primary, while all other clusters in the same regions were classified as alternative.
External validation
We used the ATAC-seq data provided by BarkBase21, mapped to CanFam3.1, and UU_Cfam_GSD_1.0/CanFam4 using the nf-core/atacseq pipeline (v1.2.2)56. Only samples from overlapping tissues (bone marrow, cardiac muscle, endocardium, liver, occipital cortex, pancreas, adenohypophysis, neurohypophysis, skeletal muscle, spleen, stomach, and thyroid glands) were used. The ChIP-seq data were obtained from Son et al. 36 and aligned to CanFam3.1 and CanFam4 using the nf-core/chipseq pipeline (v.2.0.0)57. Again, only tissue samples corresponding to our cohort were used for further analysis. The identified clusters in STRT were extended to 1000 bp upstream and 500 bp downstream and overlapped with the ATAC-seq and ChIP-seq signals to calculate enrichment.
Tissue enrichment
Similar to the method used in the Human Protein Atlas25 (HPA), we define a promoter or gene tissue as enriched if the median expression for a particular tissue is 3-fold higher than any other tissue’s mean expression. In contrast to HPA, we chose median instead of mean expression to be more robust toward outliers.
Because of the lack of annotated orthologous gene promoter regions between CanFam4 and the human reference genome, we relied on the annotated gene symbols to compare tissue-enriched genes to get 1:1 orthologues.
Promoter validation
To validate the STRT2 data, we selected five robustly expressed candidate genes with putative new first exons: ATP synthase membrane subunit e (ATP5ME), mitochondrial translational initiation factor 3 (MTIF3), troponin I3, cardiac type (TNNI3), TBC1 ___domain family member 22B (TBC1D22B) and glutamic—pyruvic transaminase (GPT). We searched RefSeq, EST alignments, and GeneID transcript models to validate the new first exons for a predicted full-length transcript sequence in CanFam3.1. We then supplied a putative first exon/5’-UTR sequence from the peak of the validation-target promoter to the proximal exon and a putative last-exon/3’-UTR sequence. We then designed primer pairs for the putative transcript using NCBI Primer-BLAST. Forward primers closest to the promoter peaks were selected (Supplementary Data 6). To validate the novel first exons, we performed RT-PCR in the tissues where the promoters of interest showed high specific expression: retina (TBC1D22B, GPT), frontal cortex (TBC1D22B, ATP5ME, GPT), testis (MTIF3), and myocardium (TNNI3). The housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was used as a positive control. Total RNA isolated from the tissues was treated with DNase I, according to the manufacturer’s instructions (MAN0012000, Thermo Fisher Scientific), to remove any possible gDNA. cDNA synthesis was performed using 1 µg of total RNA as the starting material and oligo-dT and RevertAid First Strand cDNA Synthesis Kit (K1621, Thermo Fisher Scientific) for the reverse transcription step, according to the manufacturer’s guidelines. Amplification, followed by Sanger sequencing (Finnish Institute of Molecular Medicine, Finland), was performed using a standard protocol.
Expression Atlas
To provide end-users with easy and intuitive access to the expression and tissue enrichment of DoGA promoters and genes, we generated an online Shiny app58.
The Expression panel allows observation of the expression of a certain promoter in different organ systems and tissues. A gene or promoter can be selected by name or by searching for elements in a specific region.
Tissue Enrichment panels provide access to promoters and genes enriched in specific tissues. One can select a tissue or organ system and obtain a list of the overexpressed genes. The expression of individual genes is visualized by selecting them from the list.
In all panels, one can select a comprehensive or robust dataset with a higher threshold for expression levels and replicability.
We provide the list of diseases and genes taken from OMIA and show their tissue expression in the Disease-Associated Genes tab. This includes searching by disease, gene, or disease subgroups and visualizing the expression as a scatter plot for one gene or a heatmap for multiple ones.
Expression overlapping the SNP clusters from the dog lineage and dog behavior studies is visualized as heatmaps in the SNPs tab. You can select both sets’ subsets of lineage- or behavior-associated SNPs.
You can switch between the robust and comprehensive data set for all tabs, where the comprehensive one contains all promoter regions and the robust only those with replication in at least three samples.
Whole genome sequencing
Six dogs and four wolves were whole genome sequenced (WGS) using Illumina HiSeq X ultra-high-throughput sequencing with paired-end reads (2 * 150 bp) at Novogene (UK) Company Limited and the variants identified according to the previously reported protocol and pipeline59. The WGS data has been uploaded to SRA with accession ID: PRJNA907518.
Lift-over of SNP clusters
OMIA data were downloaded as an XML file on January 12th, 2023. Data for the lineage-specific12 and the behavior-specific29 SNPs were downloaded from the publications. CanFam3.1 positions were converted to UU_Cfam_GSD_1.0/CanFam4 positions using Python 3 and the pyliftover60 module’s liftover.convert_coordinate function with the canFam3ToCanFam4 chain file (https://hgdownload.soe.ucsc.edu/goldenPath/canFam3/liftOver/canFam3ToCanFam4.over.chain.gz, accessed on January 17th 2023).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data is available through the Data Coordination Center at https://dcc.doggenomeannotation.org/doga. All analysis scripts are accessible through https://gitlab.com/doggenomeannotation/strt. All sequences of the STRT samples will be available in NCBI’s SRA BioProject PRJNA907518. Whole genome sequencing data of six dogs and four wolves have been published in the SRA under the BioProject accession number PRJNA907518, including the following Biosamples: SRR22520740, SRR22520741, SRR22520742, SRR22520743, SRR22520744, SRR22520745, SRR22520746, SRR22520747 and SRR22520748. Source data are provided with this paper.
References
Lohi, H. et al. Expanded repeat in canine epilepsy. Science 307, 81 (2005).
Lingaas, F. et al. Bayesian mixed model analysis uncovered 21 risk loci for chronic kidney disease in boxer dogs. PLOS Genet. 19, e1010599 (2023).
Hytönen, M. K. et al. Molecular characterization of three canine models of human rare bone diseases: caffey, van den ende-gupta, and raine syndromes. PLoS Genet. 12, e1006037 (2016).
Tengvall, K. et al. Bayesian model and selection signature analyses reveal risk factors for canine atopic dermatitis. Commun. Biol. 5, 1348 (2022).
Kaukonen, M. et al. A missense variant in IFT122 associated with a canine model of retinitis pigmentosa. Hum. Genet. 140, 1569–1579 (2021).
Kim, J. H. et al. Genomically complex human angiosarcoma and canine hemangiosarcoma establish convergent angiogenic transcriptional programs driven by novel gene fusions. Mol. Cancer Res. MCR 19, 847–861 (2021).
Evans, J. M. et al. Multi-omics approach identifies germline regulatory variants associated with hematopoietic malignancies in retriever dog breeds. PLoS Genet. 17, e1009543 (2021).
Rimbault, M. et al. Derived variants at six genes explain nearly half of size reduction in dog breeds. Genome Res. 23, 1985–1995 (2013).
Drögemüller, C. et al. A mutation in hirless dogs implicates FOXI3 in ectodermal development. Science 321, 1462–1462 (2008).
Brown, E. A. et al. FGF4 retrogene on CFA12 is responsible for chondrodystrophy and intervertebral disc disease in dogs. Proc. Natl. Acad. Sci. USA. 114, 11476–11481 (2017).
Meadows, J. R. S. et al. Genome sequencing of 2000 canids by the Dog10K consortium advances the understanding of demography, genome function and architecture. Genome Biol. 24, 187 (2023).
Dutrow, E. V., Serpell, J. A. & Ostrander, E. A. Domestic dog lineages reveal genetic drivers of behavioral diversification. Cell 185, 4737–4755.e18 (2022).
Sarviaho, R. et al. A novel genomic region on chromosome 11 associated with fearfulness in dogs. Transl. Psychiatry 10, 1–10 (2020).
Noh, H. J. et al. Integrating evolutionary and regulatory information with a multispecies approach implicates genes and pathways in obsessive-compulsive disorder. Nat. Commun. 8, 774 (2017).
Lindblad-Toh, K. et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819 (2005).
Hoeppner, M. P. et al. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS One 9, e91172 (2014).
Wang, C. et al. A novel canine reference genome resolves genomic architecture and uncovers transcript complexity. Commun. Biol. 4, 185 (2021).
Halo, J. V. et al. Long-read assembly of a great dane genome highlights the contribution of GC-rich sequence and mobile elements to canine genomes. Proc. Natl. Acad. Sci. USA. 118, e2016274118 (2021).
Jagannathan, V. et al. Dog10K_boxer_tasha_1.0: a long-read assembly of the dog reference genome. Genes 12, 847 (2021).
Ballard, J. W. O. et al. The Australasian dingo archetype: de novo chromosome-length genome assembly, DNA methylome, and cranial morphology. bioRxiv https://doi.org/10.1101/2023.01.26.525801 (2023).
Megquier, K. et al. BarkBase: Epigenomic annotation of canine genomes. Genes 10, 433 (2019).
van Steenbeek, F. G., Hytönen, M. K., Leegwater, P. A. J. & Lohi, H. The canine era: the rise of a biomedical model. Anim. Genet. 47, 519–527 (2016).
Adiconis, X. et al. Comprehensive comparative analysis of 5’-end RNA-sequencing methods. Nat. Methods 15, 505–511 (2018).
Fantom Consortium & others. A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Nicholas, F. W. & Hobbs, M. Online Mendelian Inheritance in Animals (OMIA). https://omia.org/ (2012).
Kaukonen, M. et al. Maternal inheritance of a recessive RBP4 effect in canine congenital eye disease. Cell Rep. 23, 2643–2652 (2018).
Deviatiiarov, R. M. et al. An atlas of transcribed human cardiac promoters and enhancers reveals an important role of regulatory elements in heart failure. Nat. Cardiovasc. Res. 2, 58–75 (2023).
Morrill, K. et al. Ancestry-inclusive dog genomics challenges popular breed stereotypes. Science 376, eabk0639 (2022).
Evans, H. E. & Christensen, G. C. Miller’s Anatomy of the Dog 3rd edn, Vol. 1130 (WB Saunders Co, 1993).
van der Spuy, J. et al. The expression of the Leber congenital amaurosis protein AIPL1 coincides with rod and cone photoreceptor development. Invest. Ophthalmol. Vis. Sci. 44, 5396–5403 (2003).
Sproll, P. et al. Assembling the jigsaw puzzle: CBX2 isoform 2 and its targets in disorders/differences of sex development. Mol. Genet. Genomic Med. 6, 785–795 (2018).
Severin, J. et al. Interactive visualization and analysis of large-scale sequencing datasets using ZENBU. Nat. Biotechnol. 32, 217–219 (2014).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Roller, M. et al. LINE retrotransposons characterize mammalian tissue-specific and evolutionarily dynamic regulatory regions. Genome Biol. 22, 62 (2021).
Son, K. H. et al. Integrative mapping of the dog epigenome: reference annotation for comparative intertissue and cross-species studies. Sci. Adv. 9, eade3399 (2023).
Bannasch, D. L. et al. Dog colour patterns explained by modular promoters of ancient canid origin. Nat. Ecol. Evol. 5, 1415–1423 (2021).
Kaukonen, M. et al. A putative silencer variant in a spontaneous canine model of retinitis pigmentosa. PLoS Genet. 16, e1008659 (2020).
Niskanen, J. E. et al. Identification of novel genetic risk factors of dilated cardiomyopathy: from canine to human. Genome Med. 15, 73 (2023).
Kirilenko, B. M. et al. Integrating gene annotation with orthology inference at scale. Science 380, eabn3107 (2023).
Islam, S. et al. Highly multiplexed and strand-specific single-cell RNA 5′ end sequencing. Nat. Protoc. 7, 813–828 (2012).
Islam, S. et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11, 163–166 (2014).
Ezer, S. et al. Generation of RNA sequencing libraries for transcriptome analysis of globin-rich tissues of the domestic dog. STAR Protoc. 2, 100995 (2021).
Feld, M. et al. The pruritus- and TH2-associated cytokine IL-31 promotes growth of sensory nerves. J. Allergy Clin. Immunol. 138, 500–508.e24 (2016).
Körber, I. et al. Gene-expression profiling suggests impaired signaling via the interferon pathway in Cstb-/- Microglia. PLoS One 11, e0158195 (2016).
Hakonen, E. et al. MANF protects human pancreatic beta cells against stress-induced cell death. Diabetologia 61, 2202–2214 (2018).
Katayama, S. et al. Delineating the healthy human skin UV response and early induction of interferon pathway in cutaneous lupus erythematosus. J. Invest. Dermatol. 139, 2058–2061.e4 (2019).
Vakkilainen, S. et al. The human long non-coding RNA gene RMRP has pleiotropic effects and regulates cell-cycle progression at G2. Sci. Rep. 9, 13758 (2019).
Katayama, S. et al. Acute wheeze-specific gene module shows correlation with vitamin D and asthma medication. Eur. Respir. J. 55, 1901330 (2020).
Koel, M. et al. Human endometrial cell-type-specific RNA sequencing provides new insights into the embryo-endometrium interplay. Hum. Reprod. Open 2022, hoac043 (2022).
Wedenoja, S. et al. Fetal HLA-G mediated immune tolerance and interferon response in preeclampsia. EBioMedicine 59, 102872 (2020).
Lauter, G. et al. Differentiation of ciliated human midbrain-derived LUHMES neurons. J. Cell Sci. 133, jcs249789 (2020).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Frith, M. C. et al. A code for transcription initiation in mammalian genomes. Genome Res. 18, 1–12 (2008).
Amezquita, R. A. et al. Orchestrating single-cell analysis with bioconductor. Nat. Methods 17, 137–145 (2020).
Patel, H. & others. nf-core/atacseq: nf-core/atacseq v1. 2.2—Iron Ossifrage. https://github.com/nf-core/atacseq/releases (2022).
Ewels, P. et al. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38, 276–278 (2022).
Chang, W. et al. Shiny: Web Application Framework for R. https://shiny.posit.co (2022).
Heinonen, T. et al. A loss-of-function variant in canine GLRA1 associates with a neurological disorder resembling human hyperekplexia. Hum. Genet. 142, 1221–1230 (2023).
Tretyakov, K. Pyliftover: Python Library for Lftover of Genomic oordinates. https://pypi.org/project/pyliftover/ (2019).
Acknowledgements
While preparing this manuscript, our dear friend and colleague Abdul Kadir Mukarram unexpectedly passed away. We have been indebted to his contributions to the DoGA project since its beginning. Furthermore, we thank all the DoGA consortium members and collaborators. We acknowledge the Institute for Molecular Medicine Finland core facility (FIMM) and the Biomedicum Functional Genomics Unit (FuGU) at the University of Helsinki for their sequencing services. The computations were performed on resources provided by SNIC through the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under project uppstore2017255 and snic2017-7-411 and on the IT Center for Science, CSC, Finland. We thank the personnel Mari Heikkilä and Heini Niinimäki from Ranua Wildlife Park, Jari Ylönen and Jouni Aspi from the University of Oulu, and Marko Haapakoski from Ähtäri Zoo for assisting with the samples and data from the Finnish wolves. We are grateful to all the dog owners who donated samples from their dogs for this study. This study was funded by the Jane and Aatos Erkko Foundation, with contributions from Wisdom Health and Kinship Co to the genomic part of the study. SK was supported by the Jane and Aatos Erkko Foundation.
Author information
Authors and Affiliations
Consortia
Contributions
Conceptualization and supervision: COD, JK, HL. Study design: COD, JK, HL, MH, MKH, AKM, MK, PS, NA, CA, IQ, AI, SE, SK. Sample collection: MKH, MK, HL, DB, OG, PS, NA, KK, IQ, AR, AS, DB, JS, DoGA. Consortium. Clinical examination: TSJ. Library preparation: SE. Data analysis and digital resources: MH, AKM, MA, JN, FM, NS, CA, IS, MY, SK, NS, AI, IT. Funding: HL, JK, COD. Manuscript drafting: MH, HL. Manuscript editing: All.
Corresponding authors
Ethics declarations
Competing interests
HL has consulted from Wisdom Health and Kinship Co in the past, who provides genetic testing services to dogs and partially funded this study. The other authors have no conflict of interests.
Peer review
Peer review information
Nature Communications thanks Je-Yoel Cho and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hörtenhuber, M., Hytönen, M.K., Mukarram, A.K. et al. The DoGA consortium expression atlas of promoters and genes in 100 canine tissues. Nat Commun 15, 9082 (2024). https://doi.org/10.1038/s41467-024-52798-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52798-1
This article is cited by
-
Evaluation of normalisation strategies for qPCR data obtained from canine gastrointestinal tissues with different pathologies
Scientific Reports (2025)
-
The RSPO2 gene is associated with bilateral anterior amelia in Chihuahuas
Mammalian Genome (2025)