Abstract
Excessive glucocorticoid (GC) action is linked to various metabolic disorders. Recent findings suggest that disrupting skeletal GC signaling prevents bone loss and alleviates metabolic disorders in high-fat diet (HFD)-fed obese mice, underpinning the neglected contribution of skeletal GC action to obesity and related bone loss. Here, we show that the elevated expression of 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1), the enzyme driving local GC activation, and GC signaling in osteoblasts, are associated with bone loss and obesity in HFD-fed male mice. Osteoblast-specific 11β-HSD1 knockout male mice exhibit resistance to HFD-induced bone loss and metabolic disorders. Mechanistically, elevated 11β-HSD1 restrains glucose uptake and osteogenic activity in osteoblast. Pharmacologically inhibiting osteoblastic 11β-HSD1 by using bone-targeted 11β-HSD1 inhibitor markedly promotes bone formation, ameliorates glucose handling and mitigated obesity in HFD-fed male mice. Taken together, our study demonstrates that osteoblastic 11β-HSD1 directly contributes to HFD-induced bone loss, glucose handling impairment and obesity.
Similar content being viewed by others
Introduction
High-fat diet (HFD) not only causes obesity1 and the related metabolic complications, e.g., insulin resistance, glucose intolerance and dyslipidemia2, but also induces substantial bone loss that increases the fracture risk in obese individuals3. Therefore, it is desirable to develop new therapeutic strategies for hitting two birds with one stone that combats HFD-induced obesity and bone loss simultaneously.
Glucocorticoids (GCs) are steroid hormones that regulate diverse physiological functions, including cardiovascular, metabolic, immune, and homeostatic activities4,5,6,7. However, exogenous (therapeutic) and endogenous GC excess are both harmful to health by leading to maladaptive conditions such as Cushing’s syndrome8,9, hypertension10, central obesity11, insulin resistance12 and osteoporosis13 that recapitulate the HFD-induced metabolic abnormalities. The local GC activity in cells and tissues is governed by two 11β-hydroxysteroid dehydrogenase isoforms (11β-HSDs), namely 11β-HSD1 and 11β-HSD214. 11β-HSD1 plays a pivotal role in converting intracellular inactive GC, i.e., cortisone in humans and 11-dehydrocorticosterone (11-DHC) in rodents, to their physiologically active forms, i.e., cortisol in humans and corticosterone in rodents, while 11β-HSD2 catalyzes the reverse reaction for inactivating GC15,16. Noteworthily, 11β-HSD1 has been recognized as a promising therapeutic target for obesity and diabetes in the past decade since the global 11β-HSD1 knockout mice were largely resistant to HFD-induced obesity and glucose handling impairment. Intriguingly, a recent study found that disrupting skeletal GC signaling through genetic overexpression of the GC-inactivating enzyme 11β-HSD2 could not only attenuate obesity and glucose handling impairment but also prevent bone loss in HFD-fed mice17. These findings underpin the potential contribution of skeletal GC action to the development of HFD-induced bone loss and obesity, wherein the role of osteoblastic 11β-HSD1 still remains unexplored.
In this study, we sought to explore the roles of skeletal 11β-HSD1 in HFD-induced bone loss and metabolic disorders. Using genetic and pharmacological approaches, our findings illustrate that the elevated 11β-HSD1 in osteoblasts contributes to HFD-induced bone loss, glucose handling impairment and obesity. This effect could be attributed to the GC-siganling-mediated suppression on Early Growth Response 2 (Egr2) in osteoblasts that restrains skeletal glucose uptake and bone formation. Therefore, targeting osteoblastic 11β-HSD1 could be a potential therapeutic strategy for addressing bone loss and glucose handling impairment in obese individuals.
Results
Increased skeletal 11β-HSD1 expression and GC signaling activation are associated with high-fat diet-induced systemic metabolic disorders and trabecular bone loss
We conducted a comprehensive analysis encompassing body mass index (BMI), blood glucose levels, and skeletal HSD11B1 (encoding 11β-HSD1) expression in 27 human participants (Fig. 1a). Interestingly, the HSD11B1 expressions in bone specimens were significantly elevated in overweight participants (BMI > 25) than in the normal weight participants (BMI < 25, Fig. 1b), and in individuals with hyperglycemia (fasting glucose >6.11 mmol/L) than those with normal blood glucose levels (fasting glucose<6.11 mmol/L, Fig. 1c). Moreover, either the normal weight participants with hyperglycemia (BMI < 25 and fasting glucose > 6.11 mmol/L), or the overweight participants with normal blood glucose levels (BMI > 25 and fasting glucose <6.11 mmol/L), or the overweight participants with hyperglycemia (BMI > 25 and fasting glucose > 6.11 mmol/L) exhibited significantly higher skeletal HSD11B1 expression compared to normal weight participants without hyperglycemia (BMI < 25 and fasting glucose < 6.11 mmol/L, Fig. 1d). Notably, the overweight participants with hyperglycemia showed the highest levels of skeletal HSD11B1 expression among these groups. These data hint at a negative association of the skeletal 11β-HSD1 with body weight and glycemia.
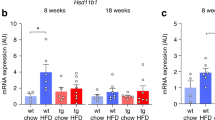
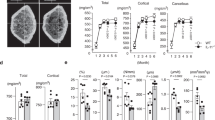
a–d The HSD11B1 expression of cancellous bone in human femoral head. a Bubble Chart depicting body mass index (BMI), blood glucose level (BG), and skeletal HSD11B1 expression in 27 human participants. b Participants with overweight (n = 8) and normal weight (n = 19). c Participants with hyperglycemia (n = 12) and normal blood glucose level (n = 15). d Participants with both normal weight and normal blood glucose level (n = 12), only hyperglycemia (n = 7), only normal glucose level (n = 4), and with both overweight and hyperglycemia (n = 4). Overweight: BMI > 25. Hyperglycemia: BG > 6.11 mmol/L. e–i Weight gain, weights of gonadal white adipose tissues (gWAT) and glucose handling tests of wild-type mice with high-fat diet (HFD) or chow diet (Chow). *P < 0.05, **P < 0.01, ***P < 0.001 when HFD vs. Chow. e Weight gain. f Weights of gWAT. g The fasting blood glucose. h The insulin resistant test (ITT). i The oral glucose tolerance test (oGTT). j–k The micro-CT analysis of wild-type mice with HFD or Chow. j The trabecular bone microstructure. k The trabecular bone volume/total volume (Tb. BV/TV) and trabecular bone density (Tb. v. BMD). l Systemic corticosterone in wild-type mice with HFD or Chow. m The mRNA expression of the 11β-HSD1 gene (Hsd11b1) and the glucocorticoid target gene Glucocorticoid-induced leucine zipper (Gilz) in liver, WAT, bone and muscle in wild-type mice with HFD, *P < 0.05, **P < 0.01 when HFD vs. Chow. n Linear regression analysis of skeletal Hsd11b1 / Gilz expression versus Tb. BV/TV, weight gain and ITT, respectively, in wild-type mice with HFD or Chow. Note: Data were presented as mean value ± SEM for (b–i, k–m). Box plot with centre line = median, cross = mean, box limits = upper and lower quartiles, whiskers = min to max (l–m). n = 4 biologically independent samples at each time-point for (e–n). Statistical significance was calculated using two-tailed Student’s t-test (a–d) and two-way ANOVA followed by two-stage step-up method by Benjamini, Krieger and Yekutieli to adjust for multiple comparisons (e–i, k–m). All tests were two-sided.
To study the alteration of endogenous GC activation and local tissue 11β-HSD1 expression in obesity, we established the diet-induced obesity model by feeding the C57BL/6 J mice with a high-fat diet (HFD, 60% kcal from fat, Figure S1a). As expected, the mice became overweight with accumulated white adipose tissue (WAT) as early as 2 weeks of HFD induction (Fig. 1e, f, Figure S1c). Meanwhile, the fasting blood glucose of HFD-fed mice was elevated compared to chow diet (Chow)-fed mice after 4-week HFD (Fig. 1g). These mice also developed insulin resistance and glucose intolerance after 4-week HFD as revealed by the insulin tolerance test (ITT) and the oral glucose tolerance test (oGTT), respectively (Fig. 1h–i, Figure S1d). Intriguingly, these mice further developed significant trabecular bone loss after 12 weeks of HFD feeding as revealed by the significant bone volume fraction (bone volume over total volume BV/TV), lower bone mineral density (v. BMD) and trabecular number (Tb. N) of distal femoral metaphysis in HFD-fed mice compared to Chow-fed mice (Fig. 1j–k, Figure S1e). No significant difference was found in cortical bone parameters in mice of these two groups (Figure S1f–g).
We then assessed both the circulating GC levels, i.e., serum corticosterone concentration and the local GC activation, i.e., the mRNA expression of Hsd11b1, the gene encoding 11β-HSD1, and Tsc22d3, the gene encoding glucocorticoid-induced leucine zipper (a GC target gene, hereafter Gilz), in the key metabolic tissues including liver, adipose tissue, bone and skeletal muscle, respectively. The serum corticosterone concentration tended to be slightly higher in HFD-fed mice than Chow-fed mice at 8 and 12 weeks of HFD induction, but no significant difference was observed at all the time points (Fig. 1l). Impressively, the expressions of Hsd11b1 and Gilz mRNAs were coincidently and markedly upregulated only in bone of mice after 8-week HFD compared to Chow-fed mice (Fig. 1m). Moreover, the protein expression levels of 11β-HSD1 and its colocalized enzyme H6PDH, which provides NADPH necessary for 11β-HSD1 to activate GC, in bone of HFD-fed mice also substantially increased from 8 to 16 weeks of feeding. But Chow-fed mice did not exhibit significant changes in these protein expression levels during the feeding period (Figure S1h). Interestingly, the expressions of Hsd11b1 mRNA rather than Gilz mRNA were slightly upregulated in skeletal muscle of HFD-fed mice at all the time points, while both mRNAs were downregulated in gonadal WAT (gWAT) of HFD-fed mice at most of the time points (Fig. 1m). But the expressions of both mRNAs in liver HFD-fed mice were not significantly altered at most of the time points except a slight upregulation of Hsd11b1 mRNA in mice with 4-week of HFD (Fig. 1m). All these findings suggest that the aberrantly increased 11β-HSD1 expression accompanied by the excessive activation of GC signaling dominantly occurs in the bone of HFD-fed mice, which follows the development of glucose handling impairment but precedes the appearance of substantial bone loss. Importantly, linear regression analysis also revealed that the skeletal Hsd11b1 and Gilz mRNA levels were both positively correlated with the weight gain, area under curve (AUC) of ITT and trabecular BV/TV, respectively, in those mice after 8-week HFD (Fig. 1n, Figure S2). Together, it suggests that increased 11β-HSD1 expression and GC signaling activation in bone are associated with HFD-induced bone loss and systemic metabolic disorders.
Osteoblast-specific Hsd11b1 knockout mice are resistant to high-fat diet-induced trabecular bone loss and systemic metabolic disorders
To explore which type of bone cells contribute to the elevated 11β-HSD1 expression and GC signaling activation in bone, we analyzed 11β-HSD1 expression within osteoblast and osteoclast of either HFD-fed or Chow-fed mice. The immunofluorescence results showed a significantly higher incidence of 11β-HSD1 in osterix- / osteocalcin-positive (Osx+/Ocn+) osteoblast lineage cells and the comparable incidence of 11β-HSD1 in osteoclast-associated receptor- /cathepsin K- positive (Oscar+/Ctsk+) osteoclast lineage cells at distal femoral metaphysis of HFD-fed mice compared to Chow-fed mice (Fig. 2a, Figure S3a). The osteoblast progenitors (Osx+Ocn-) and osteoblasts (Osx+Ocn+) were further harvested from bone marrow for RT-qPCR analysis, which showed that the Hsd11b1 mRNA expression was upregulated dominantly in osteoblasts rather than osteoblast progenitors of HFD-fed mice (Fig. 2b, Figure S3b). To determine the role of osteoblastic 11β-HSD1 in HFD-induced dysmetabolism, we established the osteoblast-specific Hsd11b1 conditional knockout mouse line (Bglap-Cre; Hsd11b1fl/fl, hereafter Ob-CKO mice) wherein the floxed allele of Hsd11b1 gene (Hsd11b1fl/fl) was excised by the osteocalcin promoter-driven Cre recombination (Bglap -Cre) (Fig. 2c, Figure S4a). The Hsd11b1 and Gilz mRNA expressions were coincidently and drastically decreased in bone rather than the other non-bone tissues of Ob-CKO mice compared to their wildtype littermates (Hsd11b1fl/fl, hereafter WT mice) (Figure S4b). Moreover, the skeletal GC conversion ratio was significantly lower in Ob-CKO mice compared to WT mice, but no significant differences in liver, fat and muscle were found between the two genotypes (Figure S4c). These data suggested that the skeletal 11β-HSD1 activity was specifically inhibited in Ob-CKO mice.
a Fluorescent immunohistochemistry analysis of 11β-HSD1 expression in bone cells of wild-type mice high-fat diet (HFD, n = 9) or chow diet (Chow, n = 9) for 16 weeks. Left: representative fluorescent images. Right: the ratio of 11β-HSD1-positive cells in either osteoblast lineage cells (Osx+ and Ocn+) or osteoclast lineage cells (Oscar+ and Ctsk+). Scale bar=50 μm. b The Hsd11b1 mRNA expression in osteoblast precursors (Osx+Ocn-) and osteoblasts (Osx+Ocn+) harvested from bone of mice with HFD (n = 5) or Chow (n = 5). c The constructing strategy. d Experimental design of osteoblast-specific Hsd11b1 knockout mice (Ob-CKO) and their littermates (WT mice) fed with HFD or Chow. e Systemic adrenocorticotropic hormone (ACTH) and corticosterone (n = 10 for Ob-CKO, n = 6 for WT). f Skeletal mRNA expression of the 11β-HSD1 gene (Hsd11b1) and the glucocorticoid target gene Glucocorticoid-induced leucine zipper (Gilz) (n = 8 for Ob-CKO, n = 6 for WT). g–j The micro-CT analysis and bone histometric analysis (n = 10 for Ob-CKO, n = 6 for WT). The data was normalized by WT-Chow group. g The trabecular bone microstructure. h The trabecular bone volume/total volume (Tb. BV/TV) and trabecular bone density (Tb. v. BMD). i The calcein double labeling. j The quantitative analysis of mineral apposition rate (MAR) and bone formation rate per bone surface (BFR/BS). k–m Weight gain, food intake and adipose tissues (n = 10 for Ob-CKO, n = 6 for WT). *P < 0.05, **P < 0.01 when HFD vs. Chow at the same genotype. k: Weight gain. The data was normalized by baseline. l: Food intake. m Weights and representative photographs of gonadal white adipose tissues (gWAT). n–p Glucose handling tests (n = 8 for Ob-CKO, n = 6 for WT). *P < 0.05, **P < 0.01, ***P < 0.001 when HFD vs. Chow at the same genotype. n Fasting blood glucose. o Insulin tolerance test (ITT). p Oral glucose tolerance test (oGTT). Note: Data were presented as mean value ± SEM for (b, e, f, h, j–p). All samples are biologically independent samples. Statistical significance was calculated using one-way ANOVA followed by Tukey’s post-hoc test (a) and two-way ANOVA followed by Sidak’s multiple comparisons test (b, e–p). All tests were two-sided.
After HFD or Chow induction (Fig. 2d), there was no significant difference in the serum concentration of either adrenocorticotropic hormone (ATCH) or corticosterone between Ob-CKO and WT mice, indicating no impact on the hypothalamic-pituitary-adrenal (HPA) axis after osteoblast-specific Hsd11b1 knockout (Fig. 2e). Subsequent results revealed that the expressions of Hsd11b1 and Gilz mRNA were coincidently upregulated in WT mice but unaltered in Ob-CKO mice when fed with HFD (Fig. 2f).
Next, we examined the bone quality and bone formation in these mice after diet induction. Interestingly, there was no significant difference in trabecular or cortical bone microarchitecture between Chow-fed Ob-CKO and WT mice (Fig. 2g, h, Figure S4d, f–g). Impressively, the trabecular microarchitecture as well as the levels of Tb. BV/TV and Tb. v. BMD in HFD-fed Ob-CKO mice resembled those in their Chow-fed controls, whereas substantial trabecular bone deterioration was presented in HFD-fed WT mice (Fig. 2g–h, Figure S4d). In addition, there was no obvious difference in the mineral apposition rate (MAR), bone formation rate per bone surface (BFR/BS), osteoblast number per bone surface (N.Ob/BS) and osteoclast number per bone surface (N.Oc/BS) at distal femoral metaphysis between the Chow-fed Ob-CKO and WT mice, respectively (Fig. 2i–j, Figure S4e). However, the MAR, BFR/BS and N.Ob/BS at distal femoral metaphysis were significantly decreased in HFD-fed WT mice compared to their Chow-fed controls, indicating reduced bone formation in WT mice after HFD feeding. Notably, the MAR, BFR/BS and N.Ob/BS all remained comparable between HFD-fed and Chow-fed Ob-CKO mice, and the values in HFD-fed Ob-CKO mice were significantly higher than those observed in HFD-fed WT mice (Fig. 2i–j, Figure S4e). Interestingly, there was no obvious change of N.Oc/BS at distal femoral metaphysis in either WT or Ob-CKO mice when fed with HFD (Figure S4e). Similarly, the mRNA expressions of bone formation-related marker genes, i.e., Runx2 (RUNX family transcription factor 2) and Bglap (bone gamma-carboxyglutamate protein, also known as osteocalcin), were comparable between HFD-fed and Chow-fed Ob-CKO mice, but significantly downregulated in HFD-fed WT mice compared to their Chow-fed controls (Figure S4h).
Interestingly, both Ob-CKO and WT mice had comparable energy intake and body weight gain when fed with Chow, whereas Ob-CKO mice exhibited similar energy intake but a notable reduction of body weight gain compared to WT mice when fed with HFD (Fig. 2k, l, Figure S4i). The HFD-induced increment of WAT weight was also attenuated in Ob-CKO mice (Fig. 2m, Figure S4j). Surprisingly, unlike WT mice, Ob-CKO mice didn’t develop obvious hyperglycemia, insulin resistance and glucose intolerance after HFD feeding (Fig. 2n–p). Furthermore, HFD feeding promoted fat deposition in bone marrow of WT mice, which was markedly compromised in Ob-CKO mice (Figure S4k). Meanwhile, as revealed by in vivo glucose uptake assay, the glucose uptake into WAT, skeletal muscle and bone were all markedly reduced in HFD-fed WT mice compared to their Chow-fed controls, whereas the skeletal glucose uptake in HFD-fed Ob-CKO mice remained comparable to their Chow-fed controls (Figure S4l). In addition, as shown in the energy expenditure analysis (Figure S4m), HFD-fed WT mice exhibited lower energy expenditure compared to Chow-fed WT mice. In contrast, Ob-CKO mice showed no significant difference in energy expenditure between those fed HFD and those fed Chow. Notably, HFD-fed Ob-CKO mice showed significantly higher energy expenditure compared to HFD-fed WT mice (Figure S4m), this increased energy expenditure in HFD-fed Ob-CKO mice likely contributes to their reduced weight gain.
Furthermore, to validate if Hsd11b1 in osteoblast dominantly contributes to HFD-induced bone loss and systemic metabolic disorders, we further compared the HFD-related phenotypes in either the osteoblast-specific (Ob-CKO), hepatocyte-specific (Alb creER; Hsd11b1fl/f, hereafter Liver-CKO) or adipocyte-specific (Adiponectin creER; Hsd11b1fl/f, hereafter Adipo-CKO) Hsd11b1 knockout mice with WT Hsd11b1fl/f mice (Figure S5a, b). To ensure uniform experimental conditions with simultaneous HFD induction, we only included male mice with the date of birth close to each other. Although the number of non-cre wild-type (WT) littermates (Hsd11b1flox/flox) obtained from each CKO strain was limited (n = 2 for Liver-CKO, n = 3 for Ob-CKO, n = 1 for Adipo-CKO), no heterogeneity of the Hsd11b1 and Gilz mRNA expression in respective tissues was found among the non-cre WT littermates of each CKO strain (Figure S5c). Therefore, we decided to pool all the WT littermates as the common WT control of all CKO strains. We further confirmed the tissue-specific knockdown of Hsd11b1 mRNA expression accompanied by the downregulated Gilz mRNA expression in each CKO strain relative to their corresponding non-cre WT littermates (Figure S5d), and the pooled non-cre wild-type littermates (Figure S5e), respectively. In addition, the mRNA expressions of Hsd11b1 and Gilz in bone were comparable between the Liver-CKO mice and WT littermates, suggesting that the skeletal Hsd11b1 expression and GC signaling were not affected in Liver-CKO mice (Figure S5e). Interestingly, the skeletal mRNA expressions of Hsd11b1 and Gilz were higher in Adipo-CKO mice compared to WT mice (Figure S5e). After HFD feeding, the body weight gain and increment of visceral WATs were found to be significantly lower, while the glucose intolerance reflected by OGTT and insulin resistance reflected by ITT were attenuated, in both Ob-CKO and Liver-CKO mice rather than Adipo-CKO mice compared to WT mice when fed with HFD (Figure S5f–j). Nevertheless, the trabecular bone microarchitecture was largely preserved in Ob-CKO mice with the highest trabecular v. BMD and BV/TV levels compared to the Liver-CKO, Adipo-CKO and WT mice, respectively, when fed with HFD (Figure S5k–m). Given the sensitivity of trabecular bone mass to slight changes in the background strains, we further assessed the ratios of Tb. v. BMD and Tb. BV/TV in each type of CKO mice relative to their respective WT littermates and found that these ratios were markedly higher in Ob-CKO mice than Liver-CKO mice, indicating that more bone mass was preserved in Ob-CKO mice as compared to Liver-CKO mice after HFD feeding. (Figure S5n). Taken together, it suggests that the elevated 11β-HSD1 was responsible for the excessive GC signaling activation in osteoblast and the impaired glucose uptake and osteoblastic bone formation in bone, thus, contributing to the HFD-induced trabecular bone loss and systemic metabolic disorders.
11β-HSD1-mediated GC signaling overactivation restrains the Egr2-governed osteogenic activity and glucose uptake in osteoblast
To explore the mechanism by which osteoblastic 11β-HSD1 regulates bone formation and glucose handling, we transfected the mouse MC3T3-E1 osteoblastic cells with a plasmid carrying the exogenous Hsd11b1 or EGFP gene to construct the 11β-HSD1-overexpressed and control osteoblastic cells (hereafter MC3T3-HSD1 and MC3T3-GFP cells), respectively (Figure S6a). RT-qPCR and western blot analysis confirmed the substantial overexpression of Hsd11b1 but little upregulation of Gilz mRNA expression in MC3T3-HSD1 cells compared to MC3T3-GFP cells without feeding with 11-DHC (the inactivated form of mouse GC) (Figure S6b–d). When fed with 11-DHC, the Gilz mRNA expression was drastically upregulated in MC3T3-HSD1 cells compared to 11-DHC-fed MC3T3-GFP cells, indicating the overactivation of GC signaling in MC3T3-HSD1 cells (Figure S6e). During osteogenic differentiation, the MC3T3-HSD1 cells exhibited decreased alkaline phosphatase (ALP) activity at 7 days after osteogenic induction and alizarin red-stained (ARS) calcium deposits at 14 and 21 days after osteogenic induction, respectively (Fig. 3a). Consistently, the mRNA expressions of Runx2 and Bglap were both notably downregulated in MC3T3-HSD1 cells during osteogenic induction (Fig. 3b). These data suggest that the elevated 11β-HSD1 over-activates GC signaling and suppresses osteogenic activity of osteoblasts.
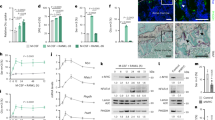
a, b Osteogenic activity of MC3T3-E1 osteoblastic cells with 11β-HSD1 overexpression (MC3T3-HSD1 cells) and transfected control cells (MC3T3-GFP cells). a Alkaline phosphatase (ALP) and alizarin red staining. b The mRNA expression of RUNX family transcription factor 2 (Runx2) and bone gamma-carboxyglutamate protein (Bglap). c, d Combination analysis of RNA sequencing (RNA-seq) and assays for transposase-accessible chromatin sequencing (ATAC-seq) in MC3T3-GFP cells and MC3T3-HSD1 cells. c Intersection of differential genes from RNA-seq and ATAC-seq. d Biological processes associated with Egr2. e The mRNA and protein expression of Egr2 in MC3T3-GFP cells and MC3T3-HSD1 cells. f The mRNA expression of Egr2 during osteogenic differentiation. g, h The osteogenic activity of MC3T3-HSD1 cells with Egr2 overexpressing. g Alizarin red staining. h Bglap mRNA expression. i The glucose uptake test of MC3T3-GFP cells and MC3T3-HSD1 cells. j, k The mRNA expression of key glucose transporter proteins and components of insulin-dependent glucose uptake pathway in MC3T3-GFP cells and MC3T3-HSD1 cells. j Glucose transporter type 1 (Glut1), 3 (Glut3) and 4 (Glut4). k: Insulin receptor (Ir), Insulin receptor substrate 1 (Irs1), Phosphatidylinositol-3-kinase catalytic subunit regulatory subunit 1 (Pik3r), alpha (Pik3ca) and subunit beta (Pik3cb). l, m The glucose uptake test and mRNA expression of Glut4 and Pik3cb in MC3T3-HSD1 cells with Egr2 overexpressing. l Glucose uptake test. m Glut4 and Pik3cb. n Dual-luciferase reporter analysis and ChIP quantitative polymerase chain reaction (ChIP-qPCR) analysis of Ege2 target interactions for Pik3cb and Glut4 gene promotors. n Pik3cb and Glut4 gene promotors. o ChIP-qPCR analysis. p Pik3cb and Glut4 gene promotors with different mutations. Note: Data were presented as mean value ± SEM for (a, b, e–m, p). n = 3 biologically independent samples for RNA-seq, ATAC-seq, western blot analysis, glucose uptake test and osteogenic staining; n = 6 biologically independent samples for RT-qPCR analysis. Statistical significance was calculated using two-tailed Student’s t-test (e), one-way ANOVA followed by Tukey’s post-hoc test (n, p), and two-way ANOVA followed by a two-stage step-up method by Benjamini, Krieger and Yekutieli (a, b, f–m) to adjust for multiple comparisons. All tests were two-sided.
We then employed RNA sequencing (RNA-seq) and transposase-accessible chromatin with sequencing (ATAC-seq, Figure S8) to examine the gene expression profile and chromatin accessibility between MC3T3-HSD1 and MC3T3-GFP cells. Combination analysis of the RNA-seq and ATAC-seq data revealed 86 dysregulated genes with accessible chromatin in their genomes, wherein 10 genes were significantly downregulated while 76 genes were significantly upregulated in MC3T3-HSD1 cells compared to MC3T3-GFP cells (Fig. 3c). Among the ten downregulated genes, we identified the Egr2, which encodes the transcriptional factor early growth factor 2, as the fourth most significantly downregulated gene with accessible chromatin in its genome. Further bioinformatic analysis showed that Egr2 was enriched in a set of genes relevant to ‘response to insulin’, ‘regulation of osteoblast differentiation’ and ‘regulation of ossification’ (Fig. 3d, Figure S7). In line with the RNA-seq data, both mRNA and protein levels of Egr2 were markedly downregulated in MC3T3-HSD1 cells compared to MC3T3-GFP cells either before or during osteogenic induction (Fig. 3e–f). A previous study documented that GC could inhibit osteocalcin transcription in osteoblasts via suppressing Egr2/Krox20-binding enhancer18. Consistently, the Egr2 mRNA expressions were notably upregulated when we silenced Gliz expression in MC3T3-HSD1 cells by transfection of small interfering RNA (siRNA) targeting Gilz (Figure S10a–c). To investigate if the GC-suppressed Egr2 expression contributes to the reduced osteogenic activity in MC3T3-HSD1 cells, we transfected the MC3T3-E1 cells with another plasmid carrying both exogenous Hsd11b1 and Egr2 genes (MC3T3-HSD1+Egr2 cells) (Figure S6f, g). As compared to the MC3T3-HSD1 cells, the MC3T3-HSD1+Egr2 cells exhibited increased calcium deposition as well as elevated Bglap, Runx2 and Osterix mRNA expressions (Fig. 3g, h, Figure S6h, i). Importantly, both the mRNA expressions of Hsd11b1 and Gilz remained upregulated in MC3T3-HSD1+Egr2 cells that resembled MC3T3-HSD1 cells when fed with 11-DHC (Figure S6g). Collectively, these data indicate that the elevated 11β-HSD1-mediated GC signaling overactivation suppresses Egr2 expression to impair osteogenic activity of osteoblasts.
Thereafter, we examined the effect of 11β-HSD1 overexpression on glucose uptake in 11-DHC-fed osteoblasts in vitro. As compared to MC3T3-GFP cells, the MC3T3-HSD1 cells showed markedly reduced glucose uptake capacity before or during osteogenic induction (Fig. 3i). Consistently, the mRNA expressions of Slc2a4 (solute carrier family 2, facilitated glucose transporter member 4), i.e., the gene encoding glucose transporter type 4 (Glut4), but not Slc2a1 and Slc2a3, i.e., the gene encoding glucose transporter type 1 and 3 (Glut1 and Glut3), were markedly downregulated in MC3T3-HSD1 cells (Fig. 3j). Then, we screened the mRNA expression of genes that encode the key pathway molecules involved in regulating the Glut4-mediated insulin-dependent glucose uptake, including insulin receptor (Ir), insulin receptor substrate (Irs), phosphoinositide-3-kinase regulatory subunit 1 (Pik3r1), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (Pik3ca) and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta (Pik3cb). Interestingly, we only identified the mRNA expressions of Pik3cb out of the other four genes that were significantly downregulated in MC3T3-HSD1 cells before or during osteogenic induction (Fig. 3k). Importantly, the Pik3cb-encoded P110β, together with the Pik3ca-encoded P110α, are the two critical subunits of phosphoinositide 3-kinase (PI3K) PI3Ks, the vital components of the PI3K/AKT signaling pathway that govern the insulin-dependent glucose uptake via regulating Glut4. Western blot analysis further verified that the protein levels of P110β and Glut4, as well as the phosphorylated AKT (pAKT) rather than the total AKT, were coincidently downregulated in MC3T3-HSD1 cells (Figure S6l). Moreover, the glucose uptake capacity was largely restored while the mRNA expressions of Pik3cb and Glut4 were markedly upregulated when we silenced Gliz expression in MC3T3-HSD1 cells. Interestingly, the MC3T3-HSD1+Egr2 cells showed enhanced glucose uptake capacity and increased mRNA expressions of Pik3cb and Glut4 either before or during osteogenic induction compared to MC3T3-HSD1 cells (Fig. 3l, m, Figure S6j, k). The promoted PI3K/AKT signaling in MC3T3-HSD1+Egr2 cells was further confirmed by the increased protein expressions of P110β, pAKT and Glut4 (Figure S6l). All these data suggest that the elevated 11β-HSD1-mediated GC signaling overactivation suppresses Egr2 expression to impair glucose uptake capacity in osteoblasts.
Next, we asked if the Egr2 directly regulates the expression of Pik3cb and Glut4. Analysis using the JASPAR online database of transcription factor binding profiles revealed a high score for Egr2 binding to the promoter region of Pik3cb and Glut4 genes (Table-S1). This observation suggests a substantial likelihood that Egr2 regulates Pik3cb and Glut4. Then, luciferase reporter assay revealed that Egr2 could directly regulate the promoter activity of both Pik3cb and Glut4 genes (Fig. 3n). In addition, chromatin immunoprecipitation (ChIP) in combination with RT-qPCR analysis confirmed that Egr2 could directly bind with the promoter of both Pik3cb and Glut4 to promote their gene expression (Fig. 3o). The binding sites at the promoter of each gene were further predicted by the open-access database of transcription factor binding profiles (JASPAR https://jaspar.elixir.no/about/), among which two of the predicted sequences with the highest binding scores were selected for gene mutation and plasmid construction followed by luciferase reporter assay (Table-S1). We found that the regulation of Egr2 on the promoter of Pik3cb and Glut4 genes was only markedly interrupted when both two binding sites of each gene promoter were simultaneously mutated (Fig. 3p), indicating that Egr2 alternatively binds with the two compensatory binding sites of each gene to regulate Pik3cb and Glut4 gene expression, respectively. Importantly, we further validated that the skeletal mRNA expressions of Egr2, Pik3cb and Glut4 were all markedly downregulated in the HFD-fed WT mice, whereas their expressions were all restored in the HFD-fed Ob-CKO mice, as compared to their Chow-fed controls (Figure S9). Collectively, all these data indicate that 11β-HSD1-mediated GC signaling overactivation restrains the Egr2-governed glucose uptake and osteogenic activity in osteoblasts.
Inhibiting 11β-HSD1 activity restores impaired osteogenic activity and glucose uptake in osteoblasts with GC signaling overactivation
Next, we sought to investigate whether inhibiting 11β-HSD1 activity could rescue the impaired osteogenic activity and glucose uptake in MC3T3-HSD1 cells by treating them with an 11β-HSD1 inhibitor (AZD8329). It was found that 11-DHC-fed MC3T3-HSD1 cells consumed a remarkably higher concentration of 11-DHC and produced a notably higher concentration of corticosterone in their culture supernatant compared to MC3T3-GFP cells, which was substantially restricted when the 11β-HSD1 inhibitor was introduced in the cell culture (Figure S10). The Gilz mRNA expression was markedly downregulated in 11-DHC-fed MC3T3-HSD1 cells with 11β-HSD1 inhibitor treatment before or during osteogenic induction (Fig. 4a, Figure S10k). Interestingly, the Hsd11b1 mRNA expression was only slightly reduced by 11β-HSD1 inhibitor during osteogenic induction (Figure S10f, k). Together, it indicates that the 11β-HSD1-mediated GC signaling overactivation in osteoblast could be efficiently blocked by 11β-HSD1 inhibitor treatment in vitro. Moreover, the 11β-HSD1 inhibitor notably rescued the downregulated expression of Egr2 in MC3T3-HSD1 cells before and during osteogenic induction (Fig. 4b–d). In addition, 11β-HSD1 inhibitor markedly promoted the calcium deposition as well as upregulated the Bglap, Runx2 and Osterix mRNA expressions in MC3T3-HSD1 cells during osteogenic induction (Fig. 4e–h). On the other hand, in line with the result of osteogenic activity assay, 11β-HSD1 inhibitor drastically enhanced glucose uptake into MC3T3-HSD1 cells and upregulated Glut4 mRNA expressions in MC3T3-HSD1 cells before or during osteogenic induction (Fig. 4i, j, Figure S10h, i). Western blot analysis further verified that the downregulated protein levels of P110β and Glut4, as well as the phosphorylated AKT (pAKT) rather than the total AKT, were coincidently upregulated in MC3T3-HSD1 cells with 11β-HSD1 inhibitor treatment (Figure S10j). Collectively, it suggests that inhibiting the 11β-HSD1 activity could restore osteogenic activity and glucose uptake in osteoblasts with GC signaling overactivation.
a The mRNA expression of glucocorticoid target gene Glucocorticoid-induced leucine zipper (Gilz) in MC3T3-GFP cells and MC3T3-HSD1 cells after treating with 11β-HSD1 inhibitor (AZD8329). b, c The mRNA and protein expression of early growth response 2 (Egr2) in MC3T3-GFP cells and MC3T3-HSD1 cells after treatment of AZD8329. b The mRNA expression. c The protein expression. d The mRNA expression of Egr2 in MC3T3-GFP cells and MC3T3-HSD1 cells after treating with AZD8329 during osteogenic differentiation. e–h The osteogenic activity of MC3T3-GFP cells and MC3T3-HSD1 cells after treatment of AZD8329. e Alizarin red staining. f Bone gamma-carboxyglutamate protein (Bglap) mRNA expression. g Runt-related transcription factor 2 (Runx2) mRNA expression. h Osterix mRNA expression. i–j The glucose uptake test and mRNA expression levels of Glucose transporter type 4 (Glut4) and Phosphatidylinositol-3-kinase catalytic subunit beta (Pik3cb) in MC3T3-GFP cells and MC3T3-HSD1 cells after treatment of AZD8329. g: Glucose uptake test. h Glut4 and Pik3cb mRNA expression. Note: Data were presented as mean value ± SEM for (a, b, d–j). n = 3 biologically independent samples for osteogenic staining, western blot analysis and LC-MS/MS analysis, n = 6 biologically independent samples for RT-qPCR analysis. Statistical significance was calculated using one-way ANOVA followed by Tukey’s post-hoc test (a–c), and two-way ANOVA followed by a two-stage step-up method by Benjamini, Krieger and Yekutieli (d–j) to adjust for multiple comparisons. All tests were two-sided.
Pharmacological inhibition of 11β-HSD1 in osteoblasts attenuates high-fat diet-induced obesity, and related glucose handling impairment and bone loss
As inspired by the protective effect of osteoblast-specific 11β-HSD1 knockout against HFD, we next sought to investigate whether pharmacological inhibition of osteoblastic 11β-HSD1 activity could combat HFD-induced dysmetabolism. To facilitate the inhibition of 11β-HSD1 activity in osteoblasts, we developed a bone-targeted 11β-HSD1 inhibitor by conjugating the AZD8329 with our previously developed bone formation surface targeting moiety (DSS)6, i.e., (DSS)6-AZD8329 (Fig. 5a, Figure S11a, b). The control 11β-HSD1 inhibitor without bone targeting specificity was also generated by conjugating AZD8329 with the control moiety (RKK)6, i.e., (RKK)6-AZD8329 (Figure S11c, d). In vitro assay confirmed that both inhibitors could significantly inhibit the conversion of 11-DHC to corticosterone in either MC3T3-HSD1 cells or MC3T3-GFP cells (Figure S12a). As revealed by IVIS imaging, there was more fluorescent signal of Cy5-labelled AZD8329 distributed in bone, but less fluorescent signal distributed in liver and adipose tissues, from mice intravenously treated with one dose of (DSS)6-AZD8329 compared to the mice with (RKK)6-AZD8329 treatment (Fig. 5b, Figure S12b). Consistently, confocal imaging showed more instances of colocalization of fluorescent signals of Cy5-labelled AZD8329 with osteocalcin-expressing osteoblasts from mice intravenously treated with one dose of (DSS)6-AZD8329 compared to the mice with (RKK)6-AZD8329 treatment (Fig. 5c, Figure S12c). Furthermore, 11β-HSD1 activity tests indicated that (DSS)6-AZD8329-treated mice showed significantly lower GC conversion ratio (Figure S12d) in bone, while (RKK)6-AZD8329-treated mice only showed slightly lower GC conversion ratio in bone, as compared to vehicle-treated mice. However, the GC conversion ratio in liver, fat and muscle were all significantly lower in (RKK)6-AZD8329-treated mice, while they were only slightly lower in (DSS)6-AZD8329-treated mice, as compared to vehicle-treated mice. These data suggest that the (DSS)6-AZD8329 was superior to (RKK)6-AZD8329 in suppressing the skeletal 11β-HSD1 activity, whereas (RKK)6-AZD8329 showed a more profound effect on suppressing 11β-HSD1 activity in non-skeletal tissues. Moreover, the data of Gilz expression in the respective tissues paralleled the above 11β-HSD1 activity trends (Figure S12e), further advocating Gilz as an indicator of local GC signaling associated with 11β-HSD1 activity. In addition, the skeletal Gilz mRNA expression was decreased by more than 50% from baseline in mice at three days after one dose of intravenous administration of (DSS)6-AZD8329, while such inhibition on Gilz mRNA expression was enhanced dose-dependently from 3 mg/kg to 10 mg/kg (Figure S12f). Therefore, we chose the 10 mg/kg dosage at a three-day interval as the regimen for the following in vivo experiments.
a Design of the bone-targeted 11β-HSD1 inhibitor, (DSS)6-AZD8329. b, c Organ distribution and skeletal distribution of (DSS)6-AZD8329 (n = 3) and (RKK)6-AZD8329 (n = 3). b The organ distribution. c The skeletal distribution, scale bar=20 μm. d Experimental design of wild-type mice treated with (DSS)6-AZD8329 and (RKK)6-AZD8329. e–g The micro-CT analysis and bone histometric analysis. e The trabecular bone and cortical bone microstructure. f The percentage change of trabecular bone volume/total volume (Tb. BV/TV) and trabecular bone density (Tb. v. BMD). g The percentage change of cortical bone density (Ct. v. BMD). h The calcein double labeling, scale bar=20 μm. i The percentage change of mineral apposition rate (MAR), bone formation rate per bone surface (BFR/BS) and the number of osteoblasts per bone surface (N.Ob/BS). j–k Weight gain and energy intake during HFD. *P < 0.05, **P < 0.01, ***P < 0.001 when other groups vs. HFD+vehicle group. j Weight gain. k Energy intake. l Weights and representative photographs of gonadal white adipose tissues (gWAT). m–o Glucose handling tests (n = 8 for two vehicle-treated groups, n = 12 for two drug-treated groups). *P < 0.05, **P < 0.01, ***P < 0.001 when other groups vs. HFD + vehicle group. m Fasting blood glucose. n: Insulin tolerance test (ITT). o Oral glucose tolerance test (oGTT). p Glucose uptake tests (n = 8 for two vehicle-treated groups, n = 12 for two drug-treated groups). Left: Representative images of glucose uptake. Right: Quantitative analysis of glucose uptake into liver, WAT, muscle and bone. q Skeletal mRNA expression of Hsd11b1, Gilz, Egr2, Pik3cb and Glut4. Note: Data were presented as mean value ± SEM for (f, g, i–q). Chow+vehicle (n = 8), mice with Chow feeding and start administration of vehicle since week 8; HFD+vehicle (n = 8), HFD + (RKK)6-AZD8329 (n = 13), HFD + (DSS)6-AZD8329 (n = 13), mice with HFD feeding and start administration of vehicle, (RKK)6-AZD8329, or (DSS)6-AZD8329, respectively, since week 8. All samples are biologically independent samples. Statistical significance was calculated using one-way ANOVA followed by Tukey’s post-hoc test (f–g, i, l, m–q) and two-way ANOVA followed by Sidak’s multiple comparisons test (j–k, n–o). All tests were two-sided.
We first tested the inhibitor in the mouse model induced by moderate HFD (43% kcal from fat, hereafter mHFD), the same HFD formula used in the previous study for testing the role of osteoblastic GC action in HFD (Figure S13a)17. Interestingly, (DSS)6-AZD8329 rather than (RKK)6-AZD8329 treatment even totally reversed the decrease of Tb. BV/TV and Tb. v. BMD, and microarchitecture deterioration (Figure S13b, c, e). In addition, (DSS)6-AZD8329 treatment resulted in significantly lower percentage decreases in MAR and N. Ob/BS as compared to (RKK)6-AZD8329 treatment (Figure S13d, e). Neither of the inhibitor treatments affected osteoclast formation (Figure S13d). Moreover, (DSS)6-AZD8329 treatment significantly alleviated body weight gain, reduced the WAT accumulation and improved glucose tolerance, while (RKK)6-AZD8329 treatment only modestly mitigated weight gain and reduced the weight of iWAT in mice with 24 weeks of mHFD (Figure S13f, h–j). Neither of the inhibitor treatments affected energy intake in mice during mHFD feeding (Figure S13g). It was noted that (DSS)6 treatment alone did not affect weight gain, adipose weight, glucose tolerance, and bone mass and microarchitecture in mHFD-fed mice, albeit they have similar mitigation on mHFD-induced bone formation reduction as (RKK)6-AZD8329 treatment. Collectively, these data suggest that pharmacological inhibition of osteoblastic 11β-HSD1 could improve glucose tolerance and promote osteoblastic bone formation to attenuate mHFD-induced obesity.
Given that the skeletal 11β-HSD1 expression became markedly upregulated in mice after 8 weeks of induction by HFD containing 60% kcal from fat in our study, we next tested the inhibitor in the mice with established obesity after 8 weeks of HFD induction (Fig. 5d). Impressively, we found that (DSS)6-AZD8329 treatment significantly mitigated the HFD-induced bone loss and microarchitecture deterioration while increasing bone formation, which is reflected by significantly lower percentage decreases in Tb. BV/TV, Tb. v. BMD, MAR and N.Ob/BS in (DSS)6-AZD8329 treatment group compared to (RKK)6-AZD8329 treatment group (Figure S14a–d, Fig. 5e–i). Neither of the inhibitor treatments affected the osteoclast formation (Figure S14c). Importantly, we found that the body weight gain was notably reduced in the HFD-fed obese mice after treatment of (DSS)6-AZD8329 and (RKK)6-AZD8329, respectively (Fig. 5j, k). Either of the inhibitor treatments markedly reduced the WAT weight, attenuated hyperglycemia and insulin resistance, and improved the glucose tolerance in HFD-fed obese mice after 8 weeks of treatment (Fig. 5l–o, Figure S14e). Impressively, as revealed in the glucose uptake assay by IVIS imaging, only (DSS)6-AZD8329 treatment significantly enhanced the glucose uptake into bone, while neither of the inhibitor treatments affected the glucose uptake into liver, WAT and skeletal muscle, respectively (Fig. 5p). Interestingly, (DSS)6-AZD8329 treatment not only significantly downregulated the skeletal mRNA expressions of Hsd11b1 and Gilz but also concurrently restored the mRNA expressions of Egr2, Pik3cb and Glut4 that were suppressed in HFD-fed mice (Fig. 5q). In contrast, (RKK)6-AZD8329 treatment only marginally inhibited the skeletal mRNA expressions of Hsd11b1 and Gilz, and slightly increased mRNA expressions of Egr2, Pik3cb, and Glut4 (Fig. 5q). Neither (DSS)6-AZD8329 nor (RKK)6-AZD8329 treatment altered the serum concentrations of ACTH and corticosterone (Figure S14f). On the other hand, we further tested the bone quality of the HFD-induced obese mice treated with Semaglutide, a glucagon-like peptide 1 receptor agonist (GLP-1RA) approved for lowering blood sugar and reducing body weight. Surprisingly, we found that Semaglutide treatment even exacerbated the HFD-induced trabecular bone loss, albeit it markedly diminished both body weight and adipose weight as well as improved glucose handling in HFD-fed mice (Figure S15). These data reflect that weight loss and metabolic improvement may not necessarily protect bone, which in turn suggests that the bone benefits of 11β-HSD1 inhibition in osteoblast could be independent of the protection from weight gain and associated metabolic benefits. Taken together, our data suggest that pharmacological inhibition of osteoblastic 11β-HSD1 could attenuate the HFD-induced obesity, and related glucose handling impairment and bone loss.
Discussion
The chronic consumption of HFD, particularly rich in saturated fat, has been associated not only with the development of obesity and metabolic syndromes in both mice and humans19,20, but also with the induction of bone loss21,22. Previous studies suggest that diet-induced obesity is accompanied by GC overproduction in key metabolic tissues17,23,24,25. Furthermore, elevated expression of 11β-HSD1, the principal regulator of local GC activation, has been observed in obese individuals14,26,27,28,29. These observations hint at the contribution of 11β-HSD1 to the HFD-induced dysmetabolism. In our study on human specimens, we observed that the 11β-HSD1 mRNA expression in bone was markedly elevated in overweight and hyperglycemic individuals. Consistently, we found that the expression of 11β-HSD1 and its colocalized enzyme H6PDH in bone were both significantly upregulated, accompanied by the elevated skeletal GC signaling in mice after 8 weeks of HFD feeding, which were closely associated with the development of HFD-induced bone loss, obesity and glucose handling impairment. Our findings, which coincide with the previous report showing the elevated 11β-HSD1 expressions and GC signaling activation in bones of mice on CD1 background fed with 18-week HFD17, challenge the preconceived notions considering liver and adipose tissues as the primary organs with elevated 11β-HSD1 and excessive GC action that majorly contribute to the adverse metabolic consequences of HFD16,30,31,32. Surprisingly, we found that the 11β-HSD1 expressions and GC signaling activation didn’t significantly elevate in the liver, WATs, and skeletal muscles of HFD-fed mice compared to Chow-fed mice. This finding aligns with a previous study reporting an early-stage reduction of 11β-HSD1 expression in the liver and WAT of HFD-fed rats, which normalized after prolonged diet induction33. Similarly, Morton et al. found that both adipose and hepatic 11β-HSD1 were downregulated after 18 weeks of HFD feeding in A/J mice and C57BL/6 J mice34. This downregulation may represent an adaptive mechanism in response to the chronic stress caused by HFD. In our study, skeletal 11β-HSD1 expression in HFD-fed mice initially decreased at the early stages of obesity but subsequently increased after 8 weeks. This biphasic response suggests that skeletal 11β-HSD1 may fail to adapt to the prolonged HFD, thereby contributing to exacerbating metabolic abnormalities, although further investigation on the dysregulation of skeletal 11β-HSD1 is needed.
We identified that the 11β-HSD1 expressions were dominantly upregulated in osteoblasts of HFD-fed mice. Previous studies have shown that global deletion of 11β-HSD1 can significantly prevent exogenous GC excess-induced and trabecular bone loss35, whereas globally transgenic overexpression of 11β-HSD1 in mice resulted in osteoporosis compared to wildtype mice36, highlighting its involvement in regulating bone metabolism. Our in vivo data of Ob-CKO mice demonstrate that conditional deletion of osteoblastic 11β-HSD1 could enhance osteoblastic bone formation and prevent HFD-induced bone loss. These findings align with the previously reported bone phenotype of mice with global35 or osteoblastic/osteocytic GC signaling blockage17,37. On the other hand, previous studies also allude to the involvement of osteoblastic GC action in regulating systemic metabolism across diverse conditions, including aging38, long-term exogenous GC administration39 and HFD17. Noteworthily, conditional deletion of osteoblastic 11β-HSD1 also protected against the HFD-induced obesity and glucose handling impairment, which recapitulates the protective phenotype of 11β-HSD1 global knockout mice with chronic high-fat feeding40,41. The Ob-CKO mice exhibited higher energy expenditure, but similar energy intake compared to WT mice after HFD feeding, which could be explained by the increased energy consumption in bone as the Ob-CKO mice exhibited improved glucose uptake dominantly in bone and enhanced osteogenic activity compared to WT mice after HFD feeding. Glucose uptake in osteoblasts is essential for osteogenesis42, while this activity reversely affects whole-body glucose metabolism because skeletal glucose uptake accounting for approximately 15% of total systemic glucose uptake43,44. Therefore, the protection from weight gain and metabolic benefits observed in our Ob-CKO mice following HFD exposure could be driven by the increased consumption of energy, e.g., glucose, in bone42. Our study substantiates that the 11β-HSD1-mediated GC signaling overactivation in osteoblasts could not only inhibit bone formation to contribute to HFD-induced bone loss, but also dominantly participate in the HFD-related adverse metabolic consequence. We also acknowledge that the skeletal 11β-HSD1 has been implicated in mediating the systemic anti-inflammatory action of GC45,46,47. In murine models of polyarthritis treated with GC, global deletion of 11β-HSD1 led to resistance against the anti-inflammatory effects of GC. This resistance was partially replicated in myeloid (including osteoclasts) but not mesenchymal (including osteoblasts) 11β-HSD1 deletion45,46. It will be worth determining the specific contribution of osteoblastic 11β-HSD1 to the systemic anti-inflammatory action of GC using our Ob-CKO model in our future study.
Noteworthily, our Liver-CKO mice rather than the Adipo-CKO mice also protected against HFD-induced obesity and glucose handling impairment. Interestingly, a previous study showed that liver-specific 11β-HSD1 knockout only mildly improved glucose tolerance without affecting fat and weight accumulation after HFD48. This discrepancy may be attributed to the different knockout strategies used in their study, i.e., Alb-Cre-mediated 11β-HSD1 knockout since embryos compared to our study, i.e., Alb-Cre ERT-mediated knockout induced before HFD feeding. Regarding the bone benefit observed in Liver-CKO mice, it could be partially explained by the metabolic improvement instead of the protection of weight gain in these mice after HFD-feeding, as the improved systemic glucose handling has been found to positively influence bone homeostasis49,50 whereas the body weight gain is primarily beneficial to bone by increasing mechanical stimulation51. It should be noted that the bone benefits in Liver-CKO mice were inferior to those observed in Ob-CKO mice, hinting the distinct underlying mechanisms that drive the bone benefits after osteoblastic and hepatic 11β-HSD1 knockout, respectively. Nevertheless, future studies in our Liver-CKO mice with enlarged sample size are still required for validation on the metabolic and bone benefits of liver-specific 11β-HSD1 knockout or inhibition.
Our mechanistic study further identified Egr2 as one of the most downregulated genes in MC3T3-HSD1 cells compared to control cells after induction with inactivated GC 11-DHC. Existing evidence indicates that Egr2 is a transcription factor directly repressed by the GC-targeted transcription factor Gliz52,53. Egr2 has been previously identified as a component of strain-sensitive pathways in osteoblasts that is responsive to anabolic signaling and mechanical loading54. It was also reported that GC could inhibit the Egr2-binding enhancer to downregulate the transcription of Bglap53. Consistently, our study demonstrated that 11β-HSD1-mediated GC signaling overactivation directly repressed Egr2 to restrain glucose uptake and osteogenic activities in osteoblasts. Further, we showed that Egr2 could directly govern the gene expression of the crucial mediators, i.e., Pik3cb and Glut4, involved in insulin-dependent glucose uptake. The Pik3cb-encoded p110β is the key component of insulin-driven PI3K-AKT pathways55,56, while Glut4 is the key glucose transporter for insulin-dependent glucose uptake57,58. Noteworthily, mice with conditional deletion of Pik3cb in osteoblast had osteopenia with reduced bone mass and impaired osteoblastic function59, while mice with conditional deletion of Glut4 in osteoblast developed peripheral insulin resistance with impaired osteoblast maturation60. Consistently, the skeletal expressions of Egr2, Pik3cb and Glut4 were also drastically downregulated in WT mice with chronic high-fat feeding, while their expression were maintained in HFD-fed Ob-CKO mice. Such evidence endorses our mechanistic findings showing that 11β-HSD1-mediated GC signaling overactivation impairs glucose uptake via repressing the Egr2-promoted Pik3cb and Glut4 expression.
Although 11β-HSD1 inhibition has long been recognized as a promising therapeutic strategy for obesity and metabolic syndrome, the clinical development of 11β-HSD1 inhibitor remains challenging and unsatisfactory61,62,63,64,65,66. Identifying the precise targeting tissues or cells might be a new direction for 11β-HSD1 inhibitor development. For instance, Liu et al. have developed an adipose tissue-targeted 11β-HSD1 inhibitor67, which exhibited improved glucose tolerance but no impact on insulin resistance. However, this inhibitor hindered glucose uptake in skeletal muscle, potentially leading to adverse effects when targeting 11β-HSD1 in adipose tissues to counteract diet-induced obesity. Our animal study suggested that targeted inhibition of osteoblastic 11β-HSD1 could be an alternative and desirable strategy for clinical translation. The peptide (DSS)6 preferentially binds to the lowly crystallized hydroxyapatite and amorphous calcium phosphonate of bone formation surface68, thus, can serve as the targeting moiety for facilitating 11β-HSD1 inhibitor approaching the osteoblasts residing at the bone formation surface in vivo68. Accordingly, we developed the bone-targeted 11β-HSD1 inhibitor (DSS)6-AZD8329 that exhibited superior osteoblast-targeting specificity than the nonspecific control inhibitor (RKK)6-AZD8329 in vivo. When evaluated in HFD-fed mice with two regimens, i.e., full course treatment during moderate HFD (43% kcal from fat) and treatment initiated when obesity was established and skeletal 11β-HSD1 was upregulated after 8 weeks of HFD (60% kcal from fat), we are encouraging to find that, in no matter which regiments, (DSS)6-AZD8329 treatment drastically improved bone formation and preserved bone quality in HFD-fed mice that was superior to (RKK)6-AZD8329 treatment. Previous clinical studies have investigated the effects of 11β-HSD1 inhibitors on bone remodeling. In clinical observation, the 11β-HSD1 inhibitor AZD4017 showed no effect on bone turnover markers in postmenopausal osteopenic women69. However, in another clinical study, AZD4017 prevented prednisolone-induced decreased bone turnover in men, suggesting that the effects of 11β-HSD1 inhibition on bone turnover may vary across different disease conditions70. (DSS)6-AZD8329 treatment could also effectively restore the skeletal glucose uptake as well as ameliorate the development of obesity and glucose handling impairment in HFD-fed mice that was comparable to or even better than (RKK)6-AZD8329 treatment. Importantly, the GLP-1RA experiment reflects that weight loss and metabolic improvement may not necessarily protect bone, which in turn suggests that the bone benefits of 11β-HSD1 inhibition in osteoblast could drive the protection from weight gain and associated metabolic benefits in Ob-CKO mice and (DSS)6-AZD8329-treated mice. It should be noted that we did not observe significant changes in HPA axis in either (DSS)6-AZD8329 or (RKK)6-AZD8329 treatment, although previous studies indicated that long-term use of 11β-HSD1 inhibitors might activate the HPA axis because of negative feedback71,72,73,74,75,76. Nonetheless, the potential side effects of long-term inhibition of osteoblastic 11β-HSD1 need to be further investigated before clinical translation.
In summary, we found that elevated 11β-HSD1 in osteoblast could mediate the GC overactivation in osteoblast to restrain skeletal glucose uptake and bone formation, thus contributing to HFD-induced bone loss, obesity, and glucose handling impairment (Fig. 6). Our study also suggests that targeting osteoblastic 11β-HSD1 could be a promising strategy for reducing body weight, improving glucose metabolism and strengthening bone in obese individuals.
In the context of HFD-induced obesity, osteoblastic 11β-HSD1 levels are upregulated, leading to overactivated glucocorticoid (GC) signaling. This augmented GC signaling downregulates the expression of Early Growth Response 2 (Egr2) in osteoblasts, and the suppressed Egr2 further restrains skeletal glucose uptake and bone formation. Consequently, the reduction in skeletal glucose uptake and bone formation contributes to bone loss and glucose handling impairment associated with obesity. This figure was created in BioRender. Zhang, G. (2023) BioRender.com/c52z877.
Methods
All animal experiments were conducted in compliance with approved protocols by the Hong Kong Baptist University animal care committees. All research complies with relevant ethical regulations.
In vivo study
Mouse housing and HFD feeding
This study was approved by the Committees of Animal Ethics and Experimental Safety of Hong Kong Baptist University (Ethical Permission NO: REC/22-23/0593 and REC/21-22/0404), demonstrating compliance with ethical standards. Bglap-Cre mice (Strain NO. 019509) were purchased from Jackson Lab. The Hsd11b1fl/fl mice (Strain NO. T008729), Alb creER mice (Strain NO. T017784) and Adiponectin creER mice (Strain NO. T052679) were purchased from GemPharmatech Co. Ltd (China).
To generate the Ob-CKO mice (Bglap-Cre; Hsd11b1fl/fl mice) required for the experiment, Hsd11b1fl/+ mice were firstly crossed with Bglap-Cre mice to produce Bglap-Cre; Hsd11b1fl/+ mice. Then, Bglap-Cre; Hsd11b1fl/+ mice were crossed with Hsd11b1fl/fl mice to generate Bglap-Cre; Hsd11b1fl/fl mice (Ob-CKO mice). These Bglap-Cre; Hsd11b1fl/fl mice were crossed with Hsd11b1fl/fl mice to generate CKO mice (Bglap-Cre; Hsd11b1fl/fl mice) and littermates (Hsd11b1fl/fl mice) for conducting experiments. Liver-CKO mice (Alb-CreER; Hsd11b1fl/fl mice) and Adipose-CKO mice (Adiponectin-CreER; Hsd11b1fl/fl mice) were also generated by crossing Hsd11b1flox/flox mice with each type of cre mice separately. The corresponding non-cre littermates (Hsd11b1fl/fl mice) generated during the breeding of each strain served as controls in the respective animal experiments.
The male mice were housed in the animal house with a 12 h light and 12 h dark cycle and were given libitum access to food and water. At 2-month old, male mice were started diet induction. The HFD (60% kcal from fat, Research Diet #D12492, 5.24 kcal/g) or Chow diet (10% kcal from fat, Research Diet #D12450J, 3.76 kcal/g) were purchased from Research Diet (NY, USA). The moderate HFD (43 % energy from fat, #SF14-144, 16.3 MJ/kg) and relative Chow diet (14% energy from fat, #SF14-008, 13.8 MJ/kg) was purchased from Specialty Feeds (WA, Australia). The weight and food intake were examined every week. Energy intake was calculated based on the caloric values obtained from Research Diets, which were 5.24 kcal/g for the HFD and 3.76 kcal/g for Chow. At designated time points, the mice were euthanized humanely for sample collection and testing.
OGTT and ITT
OGTT and ITT were performed on mice after a period of fasting. Prior to OGTT, the mice underwent a 12-hour fast and a baseline blood glucose reading was taken. Subsequently, the mice were administered orally administered with 20% glucose at 2 g/kg, and blood glucose levels were measured at intervals of 15, 30, 60, 90, and 120 min after the glucose administration. For ITT, the mice were subjected to a 4 h fast. Insulin (00169-1834-11, Novo Nordisk, Denmark) was injected with 0.75 U/kg body weight in mice, and blood glucose levels were measured at 0, 15, 30 and 60 min after the injection. Blood samples were collected from a tail prick and blood glucose levels were determined using glucose strips and an Accu-check glucometer (Roche)39. The total glucose areas under the curve (AUC) were calculated to assess the extent of glucose response. Three technical replicates were performed for each test.
Micro-computed tomography (micro-CT)
Micro-CT (VivaCT 40, SCANCO Medical AG, Switzerland) was utilized to evaluate trabecular and cortical bone of distal and midshaft femur on the left side according to the manufacturer’s instructions60,77. The femur and tibia images were reconstructed with voxel sizes of 12.5 μm, utilizing an integration time of 200 ms, 70 kVp, 114 μA, and 260 thresholds. For trabecular bone, the central region of 70% vertebral height was selected, extending from the proximal growth plate towards the vertebral body. Regions of interest (ROI) of trabecular bone from 100 consecutive layers were selected. The trabecular bone volume/total volume (Tb. BV/TV), trabecular bone density (Tb. v. BMD), thickness (Tb. Th) and number (Tb. N) were measured. The automatic threshold algorithm was employed to analyze the cortical bone of 100 slices in the distal 50% of the femur, and cortical bone parameters, such as cortical bone volume/total volume (Ct. BV/TV), bone density (Ct. BMD), porosity (Ct. porosity) and thickness (Ct. Th) were analyzed.
Serum corticosterone and ACTH measurement
Serum concentrations of mouse corticosterone (ab108821, Abcam, USA) and ACTH (ab263880, Abcam, USA) were measured by enzyme-linked immunosorbent assay (ELISA) kits in accordance with the manufacturer’s guidelines. Three technical replicates were performed for each test.
Paraffin section and H&E staining
Following sacrifice, the tibia was fixed in 4% paraformaldehyde for 48 h. Subsequently, the samples underwent complete decalcification in 10% EDTA solution. After decalcification, tissues were embedded in paraffin and sectioned into 5 μm thick slices. These sections were then stained using H&E staining working solution according to standard procedures.
Frozen section and Immunohistochemistry
Following sacrifice, the distal end of the right femur was fixed in 4% paraformaldehyde for 48 h. The specimen was then dehydrated in sucrose solutions, with increasing concentrations of 10%, 20%, and 30% sucrose in phosphate-buffered saline (PBS) for 24 h and embedded in a compound (Sakura Finetek, Japan) at the optimal cutting temperature without decalcification. Using a CryoStar NX50 (Thermo Fisher Scientific, USA), longitudinal sections of proximal region of the specimen were obtained with a thickness of 5 μm. Fluorescence immunostaining was performed on the obtained sections. After treating the blocking buffer for one hour, samples kept at a temperature of 4 °C overnight with primary antibodies specifically targeting osteocalcin (1:100; bs-4917R, Bioss, USA), osterix (1:200; ab22552, Abcam, UK), oscar (1:500; Cell Signaling; MAB1633, R&D system), CTSK (1:100; ab19027, Abcam, UK), and 11β-HSD1 (1:100; 52041, BiCell Scientific, USA). The sections were then washed and incubated with fluorescent secondary antibodies (1:1000; A78946; A-31573; A32849TR; Thermo Fisher Scientific, USA) for 45 min, then washed three times and mounted on coverslips using a Fluoroshield mounting medium containing DAPI (ab104139, Abcam, UK)78. The mouse IgG (ab37355, Abcam, UK) was used as isotype control. The dyed sections were captured utilizing a confocal laser scanning microscope (Leica, Germany). The fluorescence images were processed by Las X software (Leica).
Fluorescence active cell sorting (FACS)
Cells in femurs and tibias were first blown off with a syringe. Then, cells were washed with PBS/1% BSA and directly incubated with the first antibody for osteocalcin (1:50; bs-4917R, Bioss, USA) and stained with donkey anti-rabbit IgG-FITC (1:100; ab6717, Abcam, UK). Subsequently, cells were exposed to the primary antibody against osterix (1:50, sc-393325, Santa Cruz, USA) and then subjected to staining using donkey anti-mice IgG-APC (1:100; ab130782, Abcam, UK). FACS (BD FACSVia™ flow cytometry system, Becton Dickinson, USA) was then performed on the stained cells population60,79. The sorted cell population was used for qRT-PCR analysis. Three technical replicates were performed for each of the four biological replicates.
Bone histomorphometry
Prior to the administration of euthanasia, all mice were injected intraperitoneally with calcein at doses of 10 mg/kg on two separate occasions, 10 and 2 days before the Micro-CT scanning procedure. After Micro-CT scanning, femurs were extracted and histologically sectioned.
Additionally, modified Tartrate-resistant acid phosphatase (TRAP; 387A-1KT, Sigma, USA) staining was conducted to examine the activity of osteoclasts. Bone static histomorphometric analyses were performed to determine the osteoblast surface (Ob.N/BS) and osteoblast surface (Oc.N/BS), while bone dynamic histomorphometric analyses were conducted to determine the mineral apposition rate (MAR) and bone formation rate per bone surface (BFR/BS). These analyses were carried out using professional image analysis software (Image J, USA) following the standardized nomenclature for bone histomorphometry80. Three technical replicates were performed for each biological sample.
Agarose gel electrophoresis of genotypes
The small piece of tail of newborn mice was collected at about 3 weeks of age. The genotypes of mice were determined by PCR analyses of genomic DNA isolated from mouse tails. The floxed Hsd11b1 allele was identified with primers: upper primers, 5’-GGAGTGACACAACAGGCAACTTC-3’ and P2 (5’-AGAGACCAGACATTAGGACACCAG-3’) and lower primers, 5’-TGGTGACTGGATAAAGGGACAG-3’ and 5’-TTTCAGCTCAGCAGGTCTGTG-3’. The genotyping for Bglap Cre transgene was performed by PCR with the primers Cre F (5’-CAAATAGCCCTGGCA GATTC-3’) and Cre R (5’-TGATACAAGGGACATCTT CC-3’).
In vivo glucose uptake
The glucose uptake of mice was assessed using the XenoLight RediJect 2-DeoxyGlucosone (2-DG)-750 kit (760567, PerkinElmer, USA), in accordance with established protocol81. Briefly, the mice were fasted for 6 h, following which DG-750 was administered via the tail vein. After a period of 12 h, the ex vivo glucose uptake of liver, fat, muscle, and bone was assessed post-dissection, utilizing an In Vivo Imaging System (IVIS, PerkinElmer, USA) with excitation and emission wavelengths of Ex745 nm/Em820 nm. The obtained data were analyzed by using the Living Image software. Three technical replicates were performed for each biological sample.
Energy expenditure
Ob-CKO mice were acclimated in the Comprehensive Lab Animal Monitoring System (Columbus Instruments) for 48 h. Then, mice were monitored continuously for 24 h to measure heat generation, oxygen consumption, and carbon dioxide production82.
11β-HSD1 activity tests and LC/MS/MS analysis
For tissue sample preparation, tissues were promptly snap-frozen in liquid nitrogen and subsequently homogenized in 0.5 mL KHBR buffer for 25 min. To this homogenate, 1 mM cofactor NADPH, 10 μM 11-DHC, and 1.5 mL KHBR buffer were added. The tissue samples without adding NADPH and 11-DHC were also extracted to measure the basal level of 11-DHC and corticosterone. The mixture was then incubated in a 37 °C water bath for 2 h for liver and fat tissues, and 4 h for muscle and bone tissues. Following this incubation period, 1 mL chloroform was added to the tissue mixture, and the resulting suspension was sonicated for 15 min and centrifuged at 4000 rpm for 5 min to collect the organic layer. This extraction process was repeated twice. The resulting liquid was evaporated under a nitrogen stream, redissolved in 1 mL methanol, and prepared for LC/MS/MS analysis.
LC/MS/MS analysis was employed to quantify the concentrations of 11-DHC and corticosterone in both tissue homogenates and cell culture media from in vitro studies. Isocratic conditions were used for the separation of 11-DHC and corticosterone83. LC/MS/MS with a TurboIonSpray ionization source operating in positive ion mode at 500 °C was utilized to simultaneously monitor 11-DHC and corticosterone. Multiple reaction monitoring (MRM) techniques were applied for specific transitions: m/z 363.2-121.2 for 11-DHC and m/z 361.2-163.2 for corticosterone. Conversion ratio was normalized to tissue weight. Each biological replicate was analyzed with three technical replicates to ensure robustness and reliability of the data.
Human bone specimen collection
The cancellous bone of human femoral head was procured during orthopaedic surgery from the patients who had experienced femoral neck fractures and received artificial hip replacement at the First Affiliated Hospital of Shantou University (Ethical Permission NO: B-2024-121) and the Third Affiliated Hospital of Southern Medical University (Ethical Permission NO: 2024-ER-010). The collection of samples was conducted ethically, with clinical approval, and explicit informed consent was obtained from all participating patients.
In vitro study
Cell culture and osteogenic differentiation
The mouse pre-osteoblastic MC3T3-E1 cells were cultured in complete α-MEM (32571-036, Gibco, USA) that contained 10% fetal bovine serum (10270-106, Gibco, USA) and 1% streptomycin/penicillin (15140-122, Gibco, USA) and incubated at 37 °C with 5% CO2. The osteogenic medium used in this study was composed of completed α-MEM supplemented with 50 µg/ml L-ascorbic acid (A4544, Sigma-Aldrich, USA) and 10 mM β-glycerophosphate (ST637, Beyotime, China). Additionally, 1 μM of 11-DHC (32907, Cayman Chemical, USA) was included in the culture medium, as specified in experimental designs.
Plasmid and siRNA transfection
Plasmids expressing GFP, Hsd11b1 and Egr2 were provided by BGscience Co., LTD, and transfected into MC3T3-E cells via Lipofectamine™ 3000 Transfection Reagent (L3000015, Thermo Fisher Scientific, USA). The commercially validated siRNAs (50 nM; siNT, siGilz) were purchased from RiboBio (China) by using Lipofectamine™ 2000 Transfection Reagent (11668027, Thermo Fisher Scientific, USA). Initially, MC3T3-E1 osteoblastic cells were seeded in a culture plate and cultured with a complete medium for 24 h until they reached 80% confluency. Subsequently, the culture medium was replaced with a fresh complete medium, and the prepared complexes were combined and incubated for 15 min at ambient temperature. Then, the complex was added to the wells and culture for 48 h.
RT-qPCR
The Trizol reagent (Invitrogen, USA) was utilized to extract Total RNA from the tissues. Next, purified RNA was subjected to cDNA synthesis through a HiScript III RT SuperMix kit (R323, Vazyme, China). Real-time PCR reactions were conducted on the 7900 HT Sequence Detection System (Applied Biosystems), employing the ChamQ Universal SYBR gPCR Master Mix (Q711, Vazyme, China). The primers were synthesized by the Sangon Biotech Co., Ltd (China). The gene expression levels were measured and normalized against the endogenous control Actin. The 2−ΔΔCT method was employed to calculate the relative fold changes. The primer sequences are listed in Table S1.
Western blot
Cellular protein extraction was performed using a Tissue Protein Extraction Reagent (78510, Thermo Scientific, USA) and the protein concentration was determined using a BCA Protein Detection Kit (23225, Thermo Scientific, USA). Equal amounts of protein (30 µg) from HSD1, GFP, and MC3T3-E1 osteoblastic cells were resolved by SDS-PAGE using a 12% polyacrylamide gel and transferred onto a PVDF membrane (Millipore, USA). Following blocking with 5% nonfat milk in TBST, the membrane was incubated overnight at 4 °C with primary antibodies (1:500) against Actin (4967, Cell Signaling, USA), p110β (ab151549, abcam, UK), AKT (4691 T, Cell Signaling, USA), phosphorylated AKT (4060 T, pAKT, Cell Signaling, USA), Glut4 (21619, SAB, USA), 11β-HSD1 (AF3397, R&D system, USA), Egr2 (EPR4004, Abcam, UK), H6PDH (EPR12338, Abcam, UK). After incubation with appropriate secondary antibodies (Goat Anti-Rabbit IgG, L3012; Rabbit Anti-Goat IgG, L3042; 1:1000, SAB, USA) conjugated with horseradish peroxidase for 1 hour, the blots were developed using an ECL kit (Thermo Scientific, USA) and exposed to film. Three technical replicates were performed for each of the three biological replicates. WB results were collected using BioRad ChemiDoc XRS chemiluminescence imaging system. The protein levels were quantified by ImageJ v2.0.0 software.
ALP staining
After treatment, the cells in each experimental group were gently washed with PBS and then fixed using 4% paraformaldehyde for 30 min. Following fixation, the cells were incubated with a working solution of BCIP/NBT ALP staining kit (C3206, Beyotime, China) for an additional hour. The resultant images were captured using a camera for further analysis. The OD value was measured at a wavelength of 500 nm for quantitative analysis. Three technical replicates were performed for each of the three biological replicates.
Alizarin red staining
After fixation, the cells were treated with Alizarin red S staining solution (C0148S, Beyotime, China) and incubated for 30 min. The resulting images were acquired using a camera. The OD value was measured at a wavelength of 500 nm for quantitative analysis. Three technical replicates were performed for each of the three biological replicates.
In vitro glucose uptake
Glucose uptake analysis was carried out utilizing a colorimetric glucose uptake kit (36503, ATT Bioquest, USA) according to manufacturer’s instructions84. The experimental protocol involved washing the cells twice with Krebs-Ringer-Phosphate-HEPES buffer and incubating them in Glucose Uptake Buffer for one hour. Subsequently, the cells were subjected to insulin stimulation for 20 min. The cells were then exposed to 2-Deoxyglucose (2-DG) for 40 min. After the treatment, Acidic Lysis Buffer was utilized to lyse the cells. The resulting lysate was subjected to the addition of a 2-DG Uptake Assay working solution, followed by incubation for 2 h. Finally, the absorbance ratio at 570/610 nm was recorded using an absorbance plate reader. Three technical replicates were performed for each of the three biological replicates.
RNA-Seq
The RNA-Seq and ATAC-Seq were performed by Guangzhou Epibiotek Co., Ltd. For RNA-Seq analysis, a total of 3 µg RNA was extracted from HSD1 and Con cells treated with 11-DHC for 24 h. The VAHTS Stranded mRNA-seq Library Prep Kit for Illumina V2 (Vazyme Biotech, China) was used to prepare the libraries in accordance with the manufacturer’s instructions. The resulting sequencing reads were aligned to the mouse Ensemble genome GRCm38 with the Hisat2 aligner (version 2.1.0) and the “rna-strandness RF” parameter. The quantification of genome-mapped reads was performed using FeatureCounts (version 1.6.3). The DESeq2 R-package was used to conduct, with a significance threshold of p < 0.05 for differential expression analysis. Three technical replicates were performed for each of the three biological replicates.
ATAC-Seq
To perform ATAC-Seq analysis, Cutadapt (v2.5) was utilized for adapter trimming and sequence filtering. The resulting reads were then aligned to the mouse Ensemble genome GRCm38 using bowtie2 (v2.11.1) with default parameters. Only the paired reads that mapped uniquely were sorted using SAMtools and retained for further analysis. To generate viewable bigwig files, deepTools (v3.2.0) was employed. Genomic regions of the identified peaks were annotated using the ChIPseeker R package. Differential sites were detected from ATAC-Seq experiments using the DiffBind R package. For motif analysis, homer (v4.10.4) was used to select the ATAC peaks that were identified, then employing a significance threshold of p < 0.05 to select open chromatin regions and target genes. The Integrative Genomics Viewer (IGV) screenshots of 10 GC responded genes are shown in Figure S8. Three technical replicates were performed for each of the three biological replicates.
Combination analysis of RNA-seq and ATAC-seq
The results from both sequencing techniques were integrated to identify overlapping differentially expressed genes. Following the identification of these overlapping differentially expressed genes, pathway enrichment analysis was conducted using the KEGG pathway and GO enrichment analysis to uncover the pathways associated with these differentially expressed genes. Biological processes were annotated for the top 10 differentially expressed genes. To provide a more intuitive representation, processes related to osteogenesis and glucose metabolism are highlighted, and the complete version can be found in Figure S7.
Luciferase reporter assay
The pGL3-basic plasmids were used to construct the pGL3-Pik3cb and pGL3-Glut4 vectors by cloning DNA fragments from mouse Pik3cb and Glut4 promotors, respectively. Subsequently, Lipofectamine 3000 reagent was used to transfect MC3T3-E1 osteoblastic cells with 0.5 μg of reporter luciferase vectors, 0.5 μg of Egr-2 plasmids, and 0.1 μg of pRL-TK. After a 48 h incubation period, the cells were detected luciferase activity through Dual-Luciferase Reporter Assay System (E1910, Promega, USA). Three technical replicates were performed for each of the three biological replicates.
Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR)
293 T cells were transfected with pCDNA3.1-3*flag-tagged plasmids encoding Egr2 and control proteins. ChIP assays (Millipore, USA) were carried out following established protocols. Cells were stimulated in 10 cm dishes, fixed with 1% formaldehyde, and sonicated in PBS supplemented with a protease inhibitor. Immunoprecipitation was performed with anti-FLAG antibody-conjugated protein A agarose beads. DNA purification was carried out using the phenol/chloroform/isoamyl alcohol (Sigma, USA) method, and precipitation was done using 3 M sodium acetate, followed by elution in DEPC water. Finally, the eluted DNA (5–10 ng) was subjected to qPCR to evaluate the Pik3cb promoter and Glut4 promoter enrichment.
Statistical analysis
The study variables were presented as mean ± standard error of the mean. Unpaired tests, One-way ANOVA with Tukey’s post-hoc tests, or Two-way ANOVA with Sidak’s multiple comparisons tests were conducted to evaluate intergroup variations. Statistical analysis and linear regression analysis were performed using GraphPad Prism 8.3.0 (GraphPad Software, USA) and P < 0.05 was regarded as statistically significant68.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper. Raw data of RNA-seq and ATAC-seq in this study can be accessed through following link in NCBI websites: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE274790(RNA-seq) and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE274974(ATAC-Seq). Source data are provided with this paper.
References
Li, J. et al. An obligatory role for neurotensin in high-fat-diet-induced obesity. Nature 533, 411–415 (2016).
Hancock, C. R. et al. High-fat diets cause insulin resistance despite an increase in muscle mitochondria. Proc. Natl. Acad. Sci. 102, 7815–7820 (2008).
Patsch, J. M. et al. Increased bone resorption and impaired bone microarchitecture in short-term and extended high-fat diet–induced obesity. Metabolism 60, 243–249 (2011).
Cain, D. W. & Cidlowski, J. A. Immune regulation by glucocorticoids. Nat. Rev. Immunol. 17, 233–247 (2017).
Quax, R. A. et al. Glucocorticoid sensitivity in health and disease. Nat. Rev. Endocrinol. 9, 670–686 (2013).
Pianca, N. et al. Glucocorticoid receptor antagonization propels endogenous cardiomyocyte proliferation and cardiac regeneration. Nat. Cardiovasc. Res. 1, 617–633 (2022).
Jeanneteau, F. D. et al. BDNF and glucocorticoids regulate corticotrophin-releasing hormone (CRH) homeostasis in the hypothalamus. Proc. Natl Acad. Sci. 109, 1305–1310 (2012).
Mitchell, F. Glucocorticoid-induced Cushing syndrome increases the risk of cardiovascular events. Nat. Rev. Endocrinol. 8, 563–563 (2012).
Lodish, M. & Stratakis, C. A. A genetic and molecular update on adrenocortical causes of Cushing syndrome. Nat. Rev. Endocrinol. 12, 255–262 (2016).
Melander, O. et al. Association between a variant in the 11 beta-hydroxysteroid dehydrogenase type 2 gene and primary hypertension. J. Hum. Hypertens. 14, 819–823 (2000).
Peckett, A. J., Wright, D. C. & Riddell, M. C. The effects of glucocorticoids on adipose tissue lipid metabolism. Metabolism 60, 1500–1510 (2011).
Caratti, G. et al. Glucocorticoid activation of anti-inflammatory macrophages protects against insulin resistance. Nat. Commun. 14, 2271 (2023).
Rizzoli, R. & Biver, E. Glucocorticoid-induced osteoporosis: who to treat with what agent? Nat. Rev. Rheumatol. 11, 98–109 (2014).
Morgan, S. A. et al. 11beta-HSD1 is the major regulator of the tissue-specific effects of circulating glucocorticoid excess. Proc. Natl Acad. Sci. 111, 2482–2491 (2014).
Lindroos, J. et al. Human but Not Mouse Adipogenesis Is Critically Dependent on LMO3. Cell Metab. 18, 62–74 (2013).
Masuzaki, H. et al. A transgenic model of visceral obesity and the metabolic syndrome. Science 294, 2166–2170 (2001).
Kim, S. et al. Osteoblastic glucocorticoid signaling exacerbates high-fat-diet- induced bone loss and obesity. Bone Res 9, 40 (2021).
Gabet, Y. et al. Krox20/EGR2 deficiency accelerates cell growth and differentiation in the monocytic lineage and decreases bone mass. Blood 116, 3964–3971 (2010).
Teijeiro, A., Garrido, A., Ferre, A., Perna, C. & Djouder, N. Inhibition of the IL-17A axis in adipocytes suppresses diet-induced obesity and metabolic disorders in mice. Nat. Metab. 3, 496–512 (2021).
Ng, S. F. et al. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature 467, 963–966 (2010).
Chen, F. et al. Flaxseed oil ameliorated high-fat-diet-induced bone loss in rats by promoting osteoblastic function in rat primary osteoblasts. Nutr. Metab. 16, 71 (2019).
Urs, S., Henderson, T., Le, P., Rosen, C. J. & Liaw, L. Tissue-specific expression of Sprouty1 in mice protects against high-fat diet-induced fat accumulation, bone loss and metabolic dysfunction. Br. J. Nutr. 108, 1025–1033 (2012).
Geer, E. B., Islam, J. & Buettner, C. Mechanisms of glucocorticoid-induced insulin resistance: focus on adipose tissue function and lipid metabolism. Endocrinol. Metab. Clin. North Am. 43, 75–102 (2014).
Tsai, S. F. et al. High-fat diet-induced increases in glucocorticoids contribute to the development of non-alcoholic fatty liver disease in mice. FASEB J. 36, e22130 (2022).
Dunford, E. C. & Riddell, M. C. The Metabolic Implications of Glucocorticoids in a High-Fat Diet Setting and the Counter-Effects of Exercise. Metabolites 6, 44 (2016).
Tomlinson, N. D. et al. Absence of Cushingoid Phenotype in a Patient with Cushing’s Disease due to Defective Cortisone to Cortisol Conversion. J. Clin. Endocrinol. Metab. 87, 57–62 (2002).
Espíndola-Antunes, Daniela CEK. Adipose Tissue Expression of 11β-Hydroxysteroid Dehydrogenase Type 1 in Cushing’s Syndrome and in Obesity. Arq. Bras. Endocrinol. Metab. 51, 1397–1403 (2007).
Desbriere, R. et al. 11β-Hydroxysteroid Dehydrogenase Type 1 mRNA is Increased in Both Visceral and Subcutaneous Adipose Tissue of Obese Patients. Obesity 14, 794–798 (2012).
Baudrand, R. et al. Overexpression of 11beta-hydroxysteroid dehydrogenase type 1 in hepatic and visceral adipose tissue is associated with metabolic disorders in morbidly obese patients. Obes. Surg. 20, 77–83 (2010).
Frayn, K. N., Arner, P. & Yki-Jarvinen, H. Fatty acid metabolism in adipose tissue, muscle and liver in health and disease. Essays Biochem 42, 89–103 (2006).
Lundbom, J. Adipose tissue and liver. J. Appl Physiol. 124, 162–167 (2018).
Paterson, J. M. et al. Metabolic syndrome without obesity: Hepatic overexpression of 11β-hydroxysteroid dehydrogenase type 1 in transgenic mice. Proc. Natl Acad. Sci. 101, 7088–7093 (2004).
Drake, A. J. et al. Reduced adipose glucocorticoid reactivation and increased hepatic glucocorticoid clearance as an early adaptation to high-fat feeding in Wistar rats. Endocrinology 146, 913–919 (2005).
Morton, N. M., Ramage, L. & Seckl, J. R. Down-regulation of adipose 11beta-hydroxysteroid dehydrogenase type 1 by high-fat feeding in mice: a potential adaptive mechanism counteracting metabolic disease. Endocrinology 145, 2707–2712 (2004).
Fenton, C. G. et al. 11beta-HSD1 plays a critical role in trabecular bone loss associated with systemic glucocorticoid therapy. Arthritis Res Ther. 21, 188 (2019).
Li, H. et al. 11beta-Hydroxysteroid Dehydrogenase Type 1 Facilitates Osteoporosis by Turning on Osteoclastogenesis through Hippo Signaling. Int J. Biol. Sci. 19, 3628–3639 (2023).
Yang, J. et al. Blocking glucocorticoid signaling in osteoblasts and osteocytes prevents mechanical unloading-induced cortical bone loss. Bone 130, 115108 (2020).
Henneicke, H. et al. Skeletal glucocorticoid signalling determines leptin resistance and obesity in aging mice. Mol. Metab. 42, 101098 (2020).
Brennan-Speranza, T. C. et al. Osteoblasts mediate the adverse effects of glucocorticoids on fuel metabolism. J. Clin. Invest 122, 4172–4189 (2012).
Morton, N. M. et al. Improved lipid and lipoprotein profile, hepatic insulin sensitivity, and glucose tolerance in 11beta-hydroxysteroid dehydrogenase type 1 null mice. J. Biol. Chem. 276, 41293–41300 (2001).
Morton, N. M. et al. Novel Adipose Tissue–Mediated Resistance to Diet-Induced Visceral Obesity in 11beta-Hydroxysteroid Dehydrogenase Type 1–Deficient Mice. Diabetes 53, 931–938 (2004).
Wei, J. et al. Glucose Uptake and Runx2 Synergize to Orchestrate Osteoblast Differentiation and Bone Formation. Cell 161, 1576–1591 (2015).
Dirckx, N. et al. Vhl deletion in osteoblasts boosts cellular glycolysis and improves global glucose metabolism. J. Clin. Invest 128, 1087–1105 (2018).
Fulzele, K. et al. Insulin receptor signaling in osteoblasts regulates postnatal bone acquisition and body composition. Cell 142, 309–319 (2010).
Fenton, C. et al. Local steroid activation is a critical mediator of the anti-inflammatory actions of therapeutic glucocorticoids. Ann. Rheum. Dis. 80, 250–260 (2021).
Fenton, C. G. et al. 11beta-Hydroxysteroid Dehydrogenase Type 1 within Osteoclasts Mediates the Bone Protective Properties of Therapeutic Corticosteroids in Chronic Inflammation. Int. J. Mol. Sci. 23, 7334 (2022).
Hardy, R. S. et al. 11 Beta-hydroxysteroid dehydrogenase type 1 regulates synovitis, joint destruction, and systemic bone loss in chronic polyarthritis. J. Autoimmun. 92, 104–113 (2018).
Lavery, G. G. et al. Lack of significant metabolic abnormalities in mice with liver-specific disruption of 11beta-hydroxysteroid dehydrogenase type 1. Endocrinology 153, 3236–3248 (2012).
Ducy, P. et al. Leptin Inhibits Bone Formation through a Hypothalamic Relay: A Central Control of Bone Mass. Cell 100, 197–207 (2000).
Yang, J., Ueharu, H. & Mishina, Y. Energy metabolism: A newly emerging target of BMP signaling in bone homeostasis. Bone 138, 115467 (2020).
Gao, H. et al. Bone marrow adipoq(+) cell population controls bone mass via sclerostin in mice. Signal Transduct. Target Ther. 8, 265 (2023).
Mittelstadt, P. R. & Ashwell, J. D. Inhibition of AP-1 by the glucocorticoid-inducible protein GILZ. J. Biol. Chem. 276, 29603–29610 (2001).
Leclerc, N., Noh, T., Khokhar, A., Smith, E. & Frenkel, B. Glucocorticoids inhibit osteocalcin transcription in osteoblasts by suppressing Egr2/Krox20-binding enhancer. Arthritis Rheum. 52, 929–939 (2005).
Zaman, G. et al. Loading-related Regulation of Transcription Factor EGR2/Krox-20 in Bone Cells Is ERK1/2 Protein-mediated and Prostaglandin, Wnt Signaling Pathway-, and Insulin-like Growth Factor-I Axis-dependent. J. Biol. Chem. 287, 3946–3962 (2012).
Molinaro, A. et al. Insulin-Driven PI3K-AKT Signaling in the Hepatocyte Is Mediated by Redundant PI3Kα and PI3Kβ Activities and Is Promoted by RAS. Cell Metab. 29, 1400–1409.e1405 (2019).
He, Y. et al. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target Ther. 6, 425 (2021).
Leto, D. & Saltiel, A. R. Regulation of glucose transport by insulin: traffic control of GLUT4. Nat. Rev. Mol. Cell Biol. 13, 383–396 (2012).
Zisman, A. et al. Targeted disruption of the glucose transporter 4 selectively in muscle causes insulin resistance and glucose intolerance. Nat. Med. 6, 924–928 (2000).
Gámez, B., Rodríguez‐Carballo, E., Graupera, M., Rosa, J. L. & Ventura, F. Class I PI‐3‐Kinase Signaling Is Critical for Bone Formation Through Regulation of SMAD1 Activity in Osteoblasts. J. Bone Min. Res 31, 1617–1630 (2016).
Li, D. et al. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat. Commun. 7, 10872 (2016).
Gregory, S. et al. 11beta-hydroxysteroid dehydrogenase type 1 inhibitor use in human disease-a systematic review and narrative synthesis. Metabolism 108, 154246 (2020).
Gathercole, L. L. et al. 11beta-Hydroxysteroid dehydrogenase 1: translational and therapeutic aspects. Endocr. Rev. 34, 525–555 (2013).
Masuzaki, H. F. & Jeffrey, S. Tissue-Specific Glucocorticoid Reactivating Enzyme, 11β-Hydroxysteoid Dehydrogenase Type 1 (11β-HSD1) - A Promising Drug Target for the Treatment of Metabolic Syndrome. Curr. Drug Targets: Immune, Endocr. Metab. Disord. 3, 255–262 (2003).
Tomlinson, J. W. & Stewart, P. M. Mechanisms of disease: Selective inhibition of 11beta-hydroxysteroid dehydrogenase type 1 as a novel treatment for the metabolic syndrome. Nat. Clin. Pr. Endocrinol. 1, 92–99 (2005).
Scott, J. S., Goldberg, F. W. & Turnbull, A. V. Medicinal chemistry of inhibitors of 11beta-hydroxysteroid dehydrogenase type 1 (11beta-HSD1). J. Med Chem. 57, 4466–4486 (2014).
Chuanxin, Z. et al. Progress in 11beta-HSD1 inhibitors for the treatment of metabolic diseases: A comprehensive guide to their chemical structure diversity in drug development. Eur. J. Med Chem. 191, 112134 (2020).
Liu, J. et al. Adipose tissue-targeted 11β-hydroxysteroid dehydrogenase type 1 inhibitor protects against diet-induced obesity. Endocr. J. 58, 199–209 (2011).
Zhang, G. et al. A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nat. Med. 18, 307–314 (2012).
Abbas, A. et al. Effect of AZD4017, a Selective 11beta-HSD1 Inhibitor, on Bone Turnover Markers in Postmenopausal Osteopenia. J. Clin. Endocrinol. Metab. 107, 2026–2035 (2022).
Othonos, N. et al. 11β-HSD1 inhibition in men mitigates prednisolone-induced adverse effects in a proof-of-concept randomised double-blind placebo-controlled trial. Nat. Commun. 14, 1025 (2023).
Oda, S. et al. An Open-label Phase I/IIa Clinical Trial of 11beta-HSD1 Inhibitor for Cushing’s Syndrome and Autonomous Cortisol Secretion. J. Clin. Endocrinol. Metab. 106, e3865–e3880 (2021).
Yadav, Y. et al. Inhibition of 11beta-Hydroxysteroid dehydrogenase-1 with AZD4017 in patients with nonalcoholic steatohepatitis or nonalcoholic fatty liver disease: A randomized, double-blind, placebo-controlled, phase II study. Diabetes Obes. Metab. 24, 881–890 (2022).
Markey, K. et al. 11beta-Hydroxysteroid dehydrogenase type 1 inhibition in idiopathic intracranial hypertension: a double-blind randomized controlled trial. Brain Commun. 2, fcz050 (2020).
Rosenstock, J. et al. The 11-beta-hydroxysteroid dehydrogenase type 1 inhibitor INCB13739 improves hyperglycemia in patients with type 2 diabetes inadequately controlled by metformin monotherapy. Diabetes Care 33, 1516–1522 (2010).
Feig, P. U. et al. Effects of an 11beta-hydroxysteroid dehydrogenase type 1 inhibitor, MK-0916, in patients with type 2 diabetes mellitus and metabolic syndrome. Diabetes Obes. Metab. 13, 498–504 (2011).
Shah, S. et al. Efficacy and safety of the selective 11beta-HSD-1 inhibitors MK-0736 and MK-0916 in overweight and obese patients with hypertension. J. Am. Soc. Hypertens. 5, 166–176 (2011).
Liang, C. et al. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nat. Med 21, 288–294 (2015).
Liu, J. et al. Exosomal transfer of osteoclast-derived miRNAs to chondrocytes contributes to osteoarthritis progression. Nat. Aging 1, 368–384 (2021).
Zhao, S. et al. Wound dressings composed of copper-doped borate bioactive glass microfibers stimulate angiogenesis and heal full-thickness skin defects in a rodent model. Biomaterials 53, 379–391 (2015).
Dempster, D. W. et al. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Min. Res 28, 2–17 (2013).
Lim, S. et al. Immunoregulatory Protein B7-H3 Reprograms Glucose Metabolism in Cancer Cells by ROS-Mediated Stabilization of HIF1alpha. Cancer Res 76, 2231–2242 (2016).
Song, R. et al. Central role of E3 ubiquitin ligase MG53 in insulin resistance and metabolic disorders. Nature 494, 375–379 (2013).
Xu, R., Sang, B. C., Navre, M. & Kassel, D. B. Cell-based assay for screening 11beta-hydroxysteroid dehydrogenase inhibitors using liquid chromatography/tandem mass spectrometry detection. Rapid Commun. Mass Spectrom. 20, 1643–1647 (2006).
Rao, X. S. et al. AMPK-mediated phosphorylation enhances the auto-inhibition of TBC1D17 to promote Rab5-dependent glucose uptake. Cell Death Differ. 28, 3214–3234 (2021).
Acknowledgements
This work was supported by Theme-based Research Scheme (T12-201/20-R, A.P.L.) and General Research Fund (12106924 J.L., 12136616 J.L., 12103519 J.L., 12102120 Y.Y.Y., 12102322 Y.Y.Y., 12100921 G.Z., and 12102223 G.Z.) and Early Career Scheme (22102823 J.L.) of Research Grants Council of Hong Kong SAR. This work is also supported by 2020 Guangdong Provincial Science and Technology Innovation Strategy Special Fund (Guangdong-Hong Kong-Macau Joint Lab, No: 2020B1212030006 A.P.L.), Interdisciplinary Research Clusters Matching Scheme of Hong Kong Baptist University (RC-IRCs/17-18/02 G.Z.), Research Matching Grant Scheme of Hong Kong Baptist University (RMGS2023 _15_03 G.Z.), Inter-institutional Collaborative Research Scheme from Hong Kong Baptist University (Project No. RC-ICRS/19-20/01 G.Z.), International Science and Technology Corporation Key Program of Jiangxi Province (20232BBH80012 A.P.L.& J.L.), The National Key R&D Program of China (2018YFA0800804 G.Z.), Youth’s Project of Guangdong Basic and Applied Basic Research Fund (2022A1515110044 D.J.L.). This work is also supported by The Young Scientists Fund of the National Natural Science Foundation of China (82304378 S.F.Y.), Guangdong-Hong Kong Technology Cooperation Funding Scheme (GHP/149/21GD, 2023A0505010015 G.Z.), The National Natural Science Foundation of China (52371251 F.Z.R.), The Fundamental and Applied Fundamental Research Fund of Guangdong Province (2022B1515120082 F.Z.R.), and The National Natural Science Foundation of China (82373733 S.Z.W.).
Author information
Authors and Affiliations
Contributions
Conceptualization: G.Z., H.Z, and J.L., In vitro Investigation: C.X.Z., S.H., W.S.Y., Y.H.Z., Z.F.C., J.Q.W., J.F.Y., D.L.X., Z.M.L. In vivo Investigation: C.X.Z., N.X.L., D.J.L., S.T.M., H.W.J., H.R.Z., Z.H.L., X.X.W., H.T.L., S.X., Z.Q.W., X.H.T., Chemical synthesis and modification: S.Z.W., Clinical sample collection: L.D., G.X.H., H.R.L., Z.G.Z., Z.T.W., Maintenance and breeding of genetically engineered mouse model: Z.Z.K., S.F.Y., C.W., Data analysis: C.X.Z., N.X.L., Z.H.Y., C.L., Y.C., L.Y.W., Y.M., Y.Y.Y., and F.F.L., Funding acquisition: A.P.L., S.Z.W., F.Z.R., J.L., and G.Z., Project administration: G.Z., J.L., B.T.Z. and A.P.L., Supervision: G.Z., J.L., H.Z. and F.Z.R., Manuscript draft: C.X.Z., and J.L. Manuscript review & editing: J.L., H.Z., and G.Z.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jan Tuckermann and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhong, C., Li, N., Wang, S. et al. Targeting osteoblastic 11β-HSD1 to combat high-fat diet-induced bone loss and obesity. Nat Commun 15, 8588 (2024). https://doi.org/10.1038/s41467-024-52965-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52965-4
This article is cited by
-
Obesity: pathophysiology and therapeutic interventions
Molecular Biomedicine (2025)