Abstract
The antioxidant protein sulfiredoxin-1 (SRX) is an oncogenic factor that promotes tumor progression, but the regulatory mechanism underlying SRX degradation remains to be understood. Herein, we report that Keap1, the substrate-specific adapter of CRL3 complex, specifically binds and promotes the ubiquitin-mediated degradation of SRX at residue K61. Keap1 knockdown accumulates SRX, which in turn facilitates colorectal cancer (CRC) metastasis by activating the activator protein-1/matrix metalloproteinase 9 (AP-1/MMP9) pathway. CRC-associated Keap1 mutants within the BACK ___domain lose the capability to ubiquitinate SRX and instead promote CRC metastasis. Moreover, inactivation of Keap1 facilitates CRC tumorigenesis and metastasis in mouse models of tumor xenograft due to SRX accumulation. Clinical sample analysis reveals that Keap1 is downregulated while SRX is overexpressed in CRC, which correlates with poor prognosis. Our findings elucidate a mechanism by which CRL3Keap1 ubiquitin ligase degrades SRX to suppress CRC progression, indicating that the Keap1-SRX axis will guide the targeted therapy towards CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is one of the most common malignancies of digestive system with a rapidly rising mortality worldwide1. Distant metastasis is an important cause of treatment failure in CRC patients2. More than 50% of CRC patients have tumor cells that metastasize from the primary site to distant organs such as lung and liver, ultimately developing into metastatic colorectal cancer (mCRC)3,4,5,6. In recent years, although therapy strategies for CRC have made progress in clinical, the overall prognosis of patients remains poor due to the high rate of postsurgical recurrence and distant metastasis7. Consequently, it is urgent to clarify the molecular mechanism of CRC tumorigenesis and metastasis and identify the potential therapeutic targets.
Thiol-based antioxidants are the largest family of antioxidant molecules, mainly playing an irreplaceable role in maintaining biological redox homeostasis, in which sulfiredoxin-1 (SRX) earns a spot8,9,10. In addition to its critical function in protecting host cells from oxidative damage by reducing the hyperoxidized peroxiredoxins, emerging evidence suggests that SRX also has the ability to promote tumorigenesis and metastasis11,12. For example, SRX facilitated the progression of lung cancer by enhancing intracellular signaling cascades, such as the activator protein-1/matrix metalloproteinase 9 (AP-1/MMP9) and the mitogen-activated protein kinase (MAPK)13. SRX also promoted cervical cancer metastasis due to the stimulation of the Wnt/β-catenin signaling pathway14. Consistent with the tumor-promoting role, the SRX protein level was upregulated in multiple types of malignancies, including cervical cancer, skin cancer, and lung cancer15. All these evidences indicate that SRX functions as an oncogenic protein, nevertheless, the upstream regulatory mechanism underlying SRX overexpression in tumors remains elusive.
Keap1 is a widely studied substrate-specific adapter of the Cullin3-RING E3 ubiquitin ligase complexes (CRL3)16,17, typically consists of a BTB ___domain, a BACK ___domain, and a Kelch ___domain made up of 6 Kelch motif repeats18. Cancer-associated Keap1 mutants generally display impaired substrate-binding capacity, which disrupts the function of the entire CRL3 E3 ubiquitin complexes19. The majority of CRC-associated Keap1 mutants are within the substrate-binding Kelch ___domain20,21,22, suggesting that there is a change in binding affinity between Keap1 and its interacting partners. Keap1 targets a variety of substrates for ubiquitination and plays important biological functions, including cell proliferation, metabolism and immunity19,23,24. Multiple oncoproteins, such as IKKβ and NRF2, are ubiquitinated and degraded by Keap1, resulting in the inhibition of tumor malignant behaviors25,26. However, the impact of Keap1 on cancer by regulating redox protein substrates has not yet been fully elucidated.
Here, we show that Keap1 targets oncoprotein SRX for ubiquitination and degradation. As such, the inactivation of Keap1 induces SRX accumulation, thereby promoting CRC metastasis by activating the AP-1/MMP9 pathway. Our findings discover an anti-tumor role of the Keap1-SRX axis, suggesting that it is a negative regulator of the AP-1/MMP9 pathway and CRC progression.
Results
SRX is a substrate of Cullin-RING ligases
Cullin-RING ligases (CRLs) are multi-subunit E3 ubiquitin ligase complexes that assembled by Cullin proteins, adapter proteins alone or together with substrate-specific adapters and RING proteins (RBX1/ROC1 or SAG/ROC2), which orchestrate the degradation of specific substrates27,28. Accumulating evidence substantiates that dysfunction of CRLs promotes tumor progression by stabilizing its downstream oncogenic substrates29,30, whereas the underlying mechanisms are not fully understood. The activation of CRLs depends on the neddylation of the Cullin subunit, which is a modification process of NEDD8 covalently binding to the Cullin proteins31,32. MLN4924, a specific inhibitor of NEDD8-activating enzyme of neddylation, blocks Cullin neddylation to inhibit CRLs activation, thereby inducing the accumulation of CRLs substrates33. Therefore, we embarked to utilize MLN4924 to identify the oncoprotein targets as potential CRLs substrates and explore their regulatory mechanisms during oncogenesis and cancer progression. With a label-free quantitative proteomic strategy, we determined the upregulated proteins upon hindered neddylation with MLN4924 in SW480 cells (Supplementary Fig. 1a). Among 169 proteins that were upregulated more than two-fold after MLN4924 treatment, the expression of SRX was significantly increased 7.5-fold (Supplementary Fig. 1b). Furthermore, we found that the expression of SRX was obviously increased in a dose- and time-dependent manner upon MLN4924 treatment in four CRC cell lines (Fig. 1a, b). In line with this notion, genetically inactivating the neddylation pathway by knocking down the key enzymes of neddylation, NEDD8-activating enzyme E1 (NAE, a heterodimer consisting of NAE1 and UBA3) and NEDD8-conjugating enzyme E2 UBC12, led to SRX accumulation, thus excluding the possibility that MLN4924-induced SRX accumulation is an off-target effect (Fig. 1c). Together, these findings suggest that SRX is a potential downstream target of CRLs.
a, b SW480, HCT116, HCT15 and RKO cells were treated with MLN4924 at the indicated concentrations or time points and then subjected to western blotting analysis. The neddylation levels of Cullin1 was detected as a positive control. c SW480 cells were transfected with negative control, NAE1, UBA3, or UBC12 siRNA for 96 h and then subjected to western blotting analysis. d–g SW480 and HCT116 cells were treated with 50 μg/mL cycloheximide (CHX) along with DMSO or MLN4924 (1 μM) and then harvested for western blotting analysis. The expression of p27 was detected as a positive control. The band density of SRX was measured by densitometric analysis using ImageJ software. h–k SW480 and HCT116 cells were treated with DMSO, MG132 (20 μM), or CQ (40 μM) in combination with 50 μg/mL CHX for the indicated time points and then subjected to western blotting analysis. The expression of p27 was detected as a positive control of MG132 treatment, while the expression of LC3B-II was detected as a positive control of CQ treatment. The band density of SRX was measured by densitometric analysis using ImageJ software. l SW480 cells were transfected with control or Cullins siRNA for 96 h and then harvested for western blotting analysis. m–p SW480 and HCT116 cells were transfected with control or Cullin3 siRNA for 96 h and then treated with 50 μg/mL CHX at the indicated time points before subjecting to western blotting analysis. The band density was measured by densitometric analysis using ImageJ software. q HCT116 cells were transfected with control or Cullin3 siRNA for 96 h and followed treated with MG132 for 6 h before subjecting to immunoprecipitation with anti-SRX antibody and western blotting analysis. r HCT116 cells were treated with DMSO or MLN4924 (1 μM) for 24 h and followed with MG132 for 6 h before subjecting to immunoprecipitation with anti-SRX antibody and western blotting analysis. Representative immunoblots shown in figures were repeated three times independently with similar results. Source data are provided as a Source Data file.
To elucidate the underlying mechanism of SRX accumulation by neddylation inhibition, we first applied cycloheximide (CHX) to block protein translation and determined the SRX turnover upon MLN4924 treatment. As shown in Fig. 1d–g, pharmacological inactivation of neddylation with MLN4924 dramatically delayed the turnover and extended the half-life of SRX in both SW480 and HCT116 cells. The inhibitory effect of MLN4924 on the degradation of CRLs substrates was confirmed by p27 accumulation, a well-known substrate of CRLs (Fig. 1d, f). Since the inactivation of CRLs causes the accumulation of ubiquitination substrates, we hypothesized the degradation of SRX was regulated through the ubiquitin-proteasome pathway. As the result shown in Fig. 1h–k, CHX-induced degradation of SRX was significantly blocked in the presence of MG132, a classical proteasome inhibitor, suggesting that SRX degradation was regulated via the ubiquitin-proteasome system. Likewise, the inhibitory effect of MG132 on the degradation of ubiquitination substrates was validated by p27 accumulation (Fig. 1h, j). Meanwhile, our data showed that lysosomal inhibitor chloroquine (CQ) had no effect on the degradation of SRX, whose efficiency was confirmed by LC3B-II accumulation, indicating that SRX degradation is not regulated by the lysosomal system (Fig. 1h–k). These results demonstrate that SRX is degraded through the CRLs and ubiquitin-proteasome pathway in CRC cells.
CRL3 mediates SRX ubiquitination and degradation
Our aforementioned results indicated that the neddylation pathway regulates SRX degradation through CRLs. Next, we determined which Cullin protein was responsible for the degradation of SRX. As shown, knockdown of Cullin3, but not other Cullins, dramatically upregulated the SRX expression (Fig. 1l). Notably, the accumulation level of SRX was positively correlated with the knockdown efficiency of Cullin3 (Supplementary Fig. 1c). Besides, downregulation of ROC1, the core subunit of CRL3, significantly induced SRX accumulation (Supplementary Fig. 1d). After determining that the ablation of Cullin3 and ROC1 induced SRX accumulation, the turnover rate of SRX upon CRL3 inactivation was investigated. As depicted in Fig. 1m–p, Cullin3 knockdown significantly delayed the turnover and prolonged the half-life of SRX in the presence of CHX in SW480 and HCT116 cells. Moreover, we found that when knocking down ROC1, SRX turnover was significantly delayed and the SRX protein half-life was prolonged upon CHX treatment (Supplementary Fig. 1e–h). Finally, we detected whether CRL3 inactivation impaired the ubiquitination of SRX and found that SRX ubiquitination was obviously inhibited by Cullin3 or ROC1 knockdown as well as MLN4924 treatment in HCT116 cells (Fig. 1q, r and Supplementary Fig. 1i). Overall, these data suggest that CRL3 ubiquitin ligase targets SRX for ubiquitination and degradation in CRC cells.
Keap1 binds to and promotes the degradation of SRX
Considering that the substrate adapter proteins of CRL3 specifically recruited and targeted its certain substrates for degradation, we, therefore, investigated the exact substrate-specific adapter of CRL3 that regulated the degradation of SRX34. First, we screened a certain number of BTB ___domain-containing adapter proteins including Keap1, SPOP, KLHL22, and LZTR1, and found that downregulation of Keap1, but not other adapters, dramatically upregulated the SRX expression (Fig. 2a). We further showed that the accumulation level of SRX was associated with the degree of Keap1 knockdown (Fig. 2b). In contrast, Flag-SRX expression was suppressed by the ectopic overexpression of HA-Keap1 in a dose-dependent manner (Fig. 2c). To verify whether Keap1 interacted with SRX, co-immunoprecipitation assay was performed. As shown, endogenous Keap1 specifically pulled-down by SRX in HCT116 cells (Fig. 2d). Importantly, we performed GST pull-down assay using purified recombinant proteins of His-SRX and GST-Keap1, and found that His-SRX directly bound with GST-Keap1 in vitro, reinforcing the direct interaction between Keap1 and SRX (Fig. 2e). In line with this idea, we observed that downregulation of Keap1 significantly extended the half-life of SRX in SW480 and HCT116 cells (Fig. 2f–i). Moreover, our results revealed that Keap1 knockdown dramatically reduced the ubiquitination of SRX in HCT116 and HEK293T cells (Fig. 2j, k). These investigations demonstrate that Keap1 E3 ubiquitin ligase interacts with and governs the ubiquitinated degradation of SRX.
a, b HCT116 cells were transfected with control or CRL3 adapters siRNAs for 96 h and then harvested for western blotting analysis. HCT116 cells were transfected with control or three different Keap1 siRNA for 96 h and then harvested for western blotting analysis. c HEK293T cells were transfected with indicated concentration of HA-Keap1 plasmid and Flag-SRX plasmid and then harvested for western blotting analysis. d HCT116 cells were pretreated with MG132 for 6 h and then harvest for immunoprecipitation with anti-SRX antibody and western blotting analysis with anti-Keap1 antibody. e Recombinant proteins of GST-Keap1 (30 μg) and His-SRX (20 μg) were purified from E. coli and in vitro pull-down assay was performed as described in methods. f–i SW480 and HCT116 cells were transfected with control or Keap1 siRNA for 96 h and followed treated with 50 μg/mL CHX at the indicated time points and then harvest for western blotting analysis. The band density was measured by densitometric analysis using ImageJ software. j, k HCT116 and HEK293T cells were transfected with control or Keap1 siRNA for 96 h and followed treated with MG132 for 6 h before subjecting to immunoprecipitation with anti-SRX antibody and western blotting analysis. l Diagram of the structure and lysine mutants of SRX. m HCT116 cells were transfected with the indicated plasmids for 24 h and then harvested for western blotting analysis. n, o HCT116 cells overexpressed SRX or SRXK61R were treated with 50 μg/mL CHX for the indicated time points and then harvested for western blotting analysis. The band density was measured by densitometric analysis using ImageJ software. p HCT116 cells were transfected with indicated plasmids and then harvested for in vivo ubiquitination assay and western blotting analysis. Representative immunoblots and gels shown in figures were repeated three times independently with similar results. Source data are provided as a Source Data file.
Having established that Keap1 promotes the ubiquitination of SRX, we proceeded to identify the ubiquitin attachment sites on SRX. To this end, we constructed three K to R point mutants on SRX and determined whether Keap1 induced SRX ubiquitination through these lysine residues (Fig. 2l). Our data showed that only the SRX-K61R mutant evaded degradation by Keap1 (Fig. 2m). The co-expression of K85R or K116R mutant did not block the degradation of SRX (Fig. 2m), suggesting that K85R and K116R in SRX were largely dispensable for Keap1-mediated SRX ubiquitination. Consistent with this finding, the half-time of SRX K61R mutant was longer than that of wild-type SRX (Fig. 2n, o). Additionally, only the SRX-K61R mutant was incapable of ubiquitination by Keap1, indicating that K61 is the primary ubiquitin attachment site catalyzed by Keap1 (Fig. 2p). Together, our findings demonstrate that Keap1 promotes the degradative ubiquitination on SRX, and that K61 of SRX serves as the predominant ubiquitin attachment site.
CRC-associated Keap1 mutants in the BACK ___domain are defective in promoting SRX ubiquitination
To date, the vast majority of Keap1 mutations associated with CRC occur within its Kelch ___domain, which is responsible for selective substrate binding35. We postulated that the Kelch ___domain of Keap1 is responsible for recruiting substrates and CRC-associated Keap1 mutants within the Kelch ___domain may be defective in mediating SRX ubiquitination. Unexpectedly, our data showed that the deletion of the BACK ___domain of Keap1 (Δ179–314), but not the Kelch or BTB ___domain, severely impaired the Keap1-SRX interaction (Fig. 3a–c). Ultimately, GST pull-down assay reinforced that BACK ___domain of Keap1 directly interacted with SRX (Fig. 3d). These findings substantiate that the BACK ___domain of Keap1 is the primary binding segment for SRX.
a Schematic depiction of Keap1-FL (full length) and its deletion mutants. b, c HEK293T cells were transfected with indicated Flag-SRX and HA-Keap1 (FL) or an indicated mutant construct for 24 h and then harvested for immunoprecipitation and western blotting analysis. d Recombinant proteins of His-SRX (20 μg), GST-Keap1 (30 μg) and GST-BACK ___domain of Keap1 (30 μg) were purified from E. coli and in vitro GST pull-down assay was performed as described in the “Methods”. e The distribution of the CRC-associated Keap1 somatic mutations in the BACK ___domain of Keap1. f HEK293T cells were transfected with indicated plasmids for 24 h and then harvested for immunoprecipitation and western blotting analysis. g HCT116 cells were transfected with indicated plasmids for 24 h and then harvested for in vivo ubiquitination assay and western blotting analysis. h The distribution of the CRC-associated SRX somatic mutations. i HEK293T cells were transfected with indicated plasmids for 24 h and then harvested for western blotting analysis. Representative immunoblots and gels shown in figures were repeated three times independently with similar results. Source data are provided as a Source Data file.
Given that SRX interacted with Keap1 in a BACK ___domain-dependent manner, we further investigated whether CRC-associated Keap1 mutations in the BACK ___domain may be defective in mediating SRX ubiquitination. For this purpose, we examined cancer sequencing data deposited in the COSMIC database and identified four BACK-localized missense Keap1 mutants (A191D, L223M, C257Y and L291M) in CRC specimens (Fig. 3e). As shown, the SRX-binding ability of Keap1 mutants L223M, C257Y and L291M were markedly attenuated as compared to Keap1-WT (Fig. 3f). In accordance with this finding, we found that ectopic expression of Keap1A191D, but not Keap1L223M, C257Y or L291M mutations, obviously increased the ubiquitination of SRX (Fig. 3g). These results indicated that CRC-associated Keap1 mutants (L223M, C257Y or L291M) with the BACK ___domain exerted dominant-negative effects to deregulate SRX because of impaired SRX-binding capacity. Next, we explored whether CRC-associated SRX mutations could permit SRX to evade Keap1-mediated ubiquitination. Nevertheless, we did not find any missense SRX mutants (P77S, G86W, R101C and R101L) that escaped the degradation by Keap1 (Fig. 3h, i), implying that cancer-derived SRX mutations occur did not allow SRX to evade Keap1-mediated ubiquitination.
Keap1 knockdown-induced CRC tumorigenesis and metastasis depend on SRX accumulation
Next, we investigated the functional contribution of dysregulated Keap1-SRX axis to CRC tumorigenesis and metastasis. To this end, we first utilized the CRISPR/Cas9 system to generate SW480 and HCT116 derivative cell lines that stably downregulated the expression of Keap1 and SRX respectively or simultaneously (Fig. 4a and Supplementary Fig. 3a). Cell viability assay and clonogenic assay demonstrated that downregulation of SRX partially rescued sgKeap1 induced-cell proliferation and clonogenic survival in HCT116 cells (Fig. 4b–d). Furthermore, we found that Keap1 knockdown obviously strengthened the migration and invasion in both SW480 and HCT116 cells, while depletion of SRX caused the reverse effects (Fig. 4e, f and Supplementary Fig. 3b, c). Strikingly, the migration and invasion ability of CRC cells induced by Keap1 depletion was significantly rescued when SRX was simultaneously knocked down, indicating that downregulation of Keap1 enhanced the migration and invasion of CRC cells via blocking the degradation of SRX (Fig. 4e, f and Supplementary Fig. 3b, c).
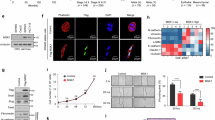
a Stable HCT116 cell lines were generated with Keap1 and SRX downregulated respectively and simultaneously, and the efficacy was validated by western blotting analysis. Cell viability in HCT116 cells with downregulated Keap1 or SRX, respectively and simultaneously, determined by ATP-Lite assay (n = 5) (b), and colony formation determined by clonogenic survival assay and representative images were shown (n = 3) (c, d). e, f Transwell assays were employed to evaluate the migration and invasion abilities of HCT116 cells in which Keap1 and SRX were downregulated respectively and simultaneously (n = 5). Representative images (e) and statistical quantifications (f), scale bar = 200 μm. g–i Stable HCT116 cell lines were subcutaneously injected into nude mice (n = 8 per group). Tumor tissues were harvested and photographed on the day of sacrifice, scale bar = 1 cm (g). Tumor size was measured with calipers at the indicated time points and converted to a tumor growth curve (h). Tumor weight was measured (i). j–m Stable HCT116 cell lines were injected into nude mice through tail vein (n = 5 per group). The tumors in lung were collected immediately after euthanized, and the number of tumor nodules was counted. Representative images, red arrow indicates the tumor nodule (j) and statistical quantifications (k). HE results of representative tumors (l) and statistical quantifications (m), scale bar = 2 mm. n, o IHC staining of lung tumor nodules using specific antibody against SRX (n = 3). Representative images (n) and statistical quantifications (o), scale bar = 50 μm. p–s Schematic diagram of orthotopic xenograft CRC mode (p). Images of visualization of the primary tumor in vivo after cellular orthotopic injection determined by bioluminescence imaging (n = 5 per group) (q). Quantification of bioluminescence imaging signals (r). Overall survival time of nude mice (s). n denotes biologically independent samples. Data were represented as mean ± SD in b, d, f, h, i, k, m, o, r. All Statistical analysis were conducted using the two-tailed unpaired Student’s t test. Representative immunoblots shown in figures were repeated three times independently with similar results. Source data are provided as a Source Data file.
To verify the in vivo role of the Keap1-SRX axis on CRC tumorigenesis and metastasis, we first established a subcutaneous-transplantation tumor model, and found that SRX depletion remarkably rescued Keap1 knockdown-induced tumor growth (Fig. 4g–i), implying that Keap1 suppressed tumor formation by promoting SRX degradation. Moreover, we constructed an experimental lung metastasis model by injecting the established CRC stable cell lines into the tail vein of mice. Compared with the control group, the number of lung metastatic nodules was significantly increased in the mice injected with Keap1-deleted cells. And the depletion of SRX reduced the number of lung metastatic nodules induced by Keap1 downregulation (Fig. 4j–m). IHC assay showed that downregulation of Keap1 resulted in a notable elevation of SRX expression in lung tumor nodules, which could be rescued by simultaneous SRX knockdown (Fig. 4n, o). Most importantly, we implemented an orthotopic transplantation mouse model to simulate the in vivo microenvironment of tumor cell growth and performed the bioluminescence imaging assay (Fig. 4p). Intriguingly, we observed that downregulation of SRX showed lower orthotopic carcinogenic and prolonged OS, which notably reversed the tumorigenicity and shorter OS upon Keap1 knockdown (Fig. 4q–s). Taken together, these in vivo results support the notion that Keap1-SRX axis silencing drives CRC progression.
Keap1-SRX axis modulates CRC metastasis through the AP-1/MMP9 pathway
Emerging evidence has shown that AP-1 is a crucial factor in SRX-mediated tumor metastasis13. To this end, we explored the involvement of AP-1 in Keap1-SRX axis regulated CRC metastasis. First, we examined the activation state of AP-1 in two different CRC cell lines. As shown, downregulation of SRX obviously attenuated the phosphorylation levels of c-Jun (the main active component of AP-1) and its upstream factor c-Jun N-terminal kinase (JNK) in both SW480 and HCT116 cells, which could reverse the AP-1 activation induced by Keap1 knockdown (Fig. 5a, b). It has been reported that the activation of AP-1 stimulated its downstream target MMP9 and promoted epithelial-mesenchymal transformation (EMT) and tumor metastasis13. We therefore sought to investigate the involvement of MMP9 and EMT in the CRC metastasis regulated by the Keap1-SRX axis. We determined the expression of MMP9 and a panel of EMT markers and found that the upregulation of MMP9 and snail, as well as the downregulation of E-cadherin when Keap1 knocked down, which could be rescued by simultaneously SRX depletion (Fig. 5a, b). Gelatin zymography assay demonstrated that SRX knockdown observably weaken the activation of MMP9 elicited by Keap1 silencing (Fig. 5c, d). These above results indicate that Keap1 knockdown leads to SRX accumulation to activate the AP-1/MMP9 pathway.
a, b The expression levels of indicated proteins in SW480 and HCT116 cells with downregulated Keap1 or SRX, respectively and simultaneously, determined by western blotting analysis. c, d MMP9 activity in the culture medium of SW480 and HCT116 cells with downregulated Keap1 or SRX, respectively and simultaneously, measured by gelatin zymography. e–g Transwell assays were employed to evaluate the migration and invasion abilities of Keap1-knockdown HCT116 cells treated with 10 μM AP-1 inhibitor T-5224 for 48 h (n = 5). The cells were harvested for western blotting analysis (e). Representative images (f) and statistical quantifications (g), scale bar = 200 μm. h–j Transwell assays were employed to evaluate the migration and invasion abilities of Keap1-knockdown HCT116 cells treated with 40 μM MMP9 inhibitor MMP-9-IN-1 for 48 h (n = 5). The cells were harvested for western blotting analysis (h). Representative images (i) and statistical quantifications (j), scale bar = 200 μm. k–n Keap1-knockdown HCT116 cells stably overexpressing EV, Flag-Keap1-WT or mutants A191D, L223M, C257Y, and L291M. Transwell assays were employed to evaluate the migration and invasion abilities of Keap1-knockdown HCT116 cells, in which Keap1-WT and indicated mutants were stably overexpressed (n = 5). Representative images (k) and statistical quantifications (l, m), scale bar = 200 μm. The cells were harvested for western blotting analysis (n). n denotes biologically independent samples. Data were represented as mean ± SD in g, j, l, m. All Statistical analysis were conducted using the two-tailed unpaired Student’s t test. n.s. denotes not significant. Representative immunoblots and gels shown in figures were repeated three times independently with similar results. Source data are provided as a Source Data file.
Furthermore, we assessed whether inhibition of AP-1/MMP9 pathway could weaken CRC metastasis upon Keap1 knockdown. As shown, AP-1 inhibitor T-5224 or MMP9 inhibitor MMP-9-IN-1 dramatically disrupted the Keap1 knockdown-induced migration and invasion of HCT116 cells and SW480 cells (Fig. 5e–j and Supplementary Fig. 4a–f), implying that inactivation of Keap1 induced CRC metastasis dependent on the AP-1/MMP9 pathway. Collectively, these data reveal that the downregulation of Keap1 enriches SRX, resulting in the metastasis of CRC cells by activating the AP-1/MMP9 pathway.
To determine whether overexpression of CRC-derived Keap1 mutants could rescue the effect of Keap1 knockdown, we re-introduced Keap1-WT and these mutants in sgKeap1 cells. We found that stably overexpressing WT and A191D mutant reversed sgKeap1-induced Srx accumulation and migration and invasion of HCT116 and SW480 cells via the AP-1/MMP9 pathway, but L223M, C257Y or L291M mutants did not reverse these effects (Fig. 5k–n and Supplementary Fig. 4g–j). Together with our aforementioned results (Fig. 3f, g), our findings collectively indicate that CRC-associated Keap1 mutants (L223M, C257Y or L291M) lose the capacity to ubiquitinate SRX and instead promote the migration and invasion of CRC cells.
Keap1 and SRX are negatively correlated in colorectal cancer tissues
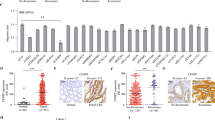
To investigate the clinical relevance of SRX in CRC, we analyzed the SRX expression levels in 128 normal tissues and 136 CRC tissues by IHC staining. Based on staining intensity, we further classified the samples into four groups with increasing staining intensity from the weakest (+) to the strongest (++++) (Fig. 6a). As summarized in Fig. 6b, SRX staining in normal tissues was weak with 97% (124/128) samples in + group. In contrast, SRX staining was elevated in CRC tissues, with 88% (120/136) in ++ to ++++ groups. By histological evaluation, we confirmed that SRX expression in CRC tissues of patients was significantly higher than that in the normal tissues (Fig. 6c). Advanced Kaplan-Meier analysis indicated that high expression of SRX was associated with worse overall survival in patients with CRC (p = 0.006, log-rank test, Fig. 6d). More strikingly, we found that CRC patients were more prone to lymph node metastasis with a gradual increase in SRX expression (Fig. 6e). Spearman’s rank correlation coefficient analysis indicated that SRX expression positively correlated with the stage of lymph node metastasis (R = 0.4348, p = 3.62E−07, Fig. 6f), strongly supporting that the upregulated SRX was prone to promote metastasis in CRC patients. Beyond that, we investigated the clinical relevance and significance of SRX in CRC patients. Notably, SRX was determined to be overexpressed in human CRC and positively correlated with the advanced stages and patients’ poor clinical outcome, as evidenced by the significant correlations with TNM (R = 0.3978, p = 1.62E−06), as well as vascular or neural invasion (R = 0.2755, p = 0.0072) (Table 1). Altogether, these findings indicate that the upregulated SRX in tumor predicts a poor prognosis in patients with CRC.
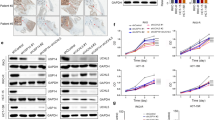
a CRC tissue arrays were stained for SRX expression. Stained tissue samples were classified into four groups (+ to ++++) according to the staining intensity. IHC staining of human CRC tissue arrays using specific antibody against SRX, scale bar = 400 μm. b The percentage of each staining group in normal or tumor tissue. c GraphPad Prism software was used to quantitatively analyze the staining intensity of SRX in normal or tumor tissue of CRC patients. d The Kaplan–Meier curves showed the overall survival rate of CRC patients according to the expression level of SRX (log-rank test). e The correlation between SRX expression and lymph node metastasis stage (N0, N1 and N2) in CRC patients (n = 126). f Correlation analysis between the expression of SRX and the stage of lymph node metastasis in CRC patients. g–i Ten paired CRC tissues with adjacent tissues and were lysed and subjected to western blotting analysis. Representative images (g) and statistical quantifications (h, i). T Tumor tissue, A Adjacent tissue. j, k Kaplan–Meier Plotter database analysis estimated the correlation between Keap1 mRNA levels and the overall survival (OS) and relapse-free survival (RFS) of CRC patients. n denotes biologically independent samples. Data were represented as mean ± SD in (c). Data were represented as symbols and lines in (h, i). Two-tailed Spearman’s correlation test was used in (f). Two-tailed paired Student’s t test was used in (h, i). Representative immunoblots shown in figures were repeated three times independently with similar results. Source data are provided as a Source Data file.
To further determine the potential role of Keap1-mediated SRX degradation during carcinogenesis, the expression of Keap1 and SRX in tumor tissues and adjacent tissues were compared. The inverse relationship between Keap1 and SRX in 10 paired CRC tissues with adjacent tissues was first confirmed by western blotting (Fig. 6g). Correlation analysis revealed that Keap1 was downregulated and strikingly correlated with high expression of SRX in CRC tissues (Fig. 6h, i). In addition, we used the Kaplan–Meier Plotter database (https://kmplot.com/analysis) to analyze the correlation between Keap1 mRNA levels and the survival prognosis of CRC patients. The results indicated that low expression of Keap1 was associated with worse overall survival (OS) (p = 0.0077) and worse relapse-free survival (RFS) (p = 0.00045) in CRC patients (Fig. 6j, k). To sum up, the above results demonstrate the negative regulatory effect of Keap1 on SRX expression in clinical outcomes of CRC patients.
Discussion
CRC is the second leading cause of cancer-related death worldwide36, but effective therapeutic strategies are still lacking. In this study, we presented evidence that oncogenic SRX was degraded by CRL3Keap1 ubiquitin ligase and revealed the role of Keap1-SRX axis in impairing CRC progression in an AP-1/MMP9 pathway-dependent manner (Fig. 7). More importantly, we found that CRC-associated Keap1 mutants within the BACK ___domain functioned as dominant-negative variants to deregulate SRX and instead facilitate CRC metastasis. Clinically, inactivation of Keap1 caused significant upregulation of SRX, and a high level of SRX was associated with poor clinical outcomes in CRC patients. Our findings uncover a mechanism of CRC tumorigenesis and metastasis controlled by the Keap1-SRX axis, implicating that targeting Keap1-SRX as an attractive strategy to anti-CRC therapy.
Apart from the involvement in regulating redox homeostasis, SRX assumes a critical role during carcinogenesis, encompassing aspects like tumor cell proliferation, migration and invasion11,37,38. SRX is preferentially overexpressed in poorly differentiated tumor cells, such as prostate cancer38, cervical cancer14 and CRC11, nevertheless, the upstream mechanism regulating SRX protein expression and its pathological contribution to tumorigenesis remain largely elusive. Abundant evidence indicated that SRX was regulated at the transcriptional level by various factors, such as chemotherapy drugs, oxidative stress-responsive transcription factors (e.g., AP-1 and NRF2), etc39. However, whether SRX could be modified at the posttranscriptional level remained insufficiently studied. In the present study, we discovered that Keap1 targeted SRX for ubiquitination and degradation, as supported by the following evidences: (1) Keap1 specifically interacted with SRX, (2) downregulation of Keap1 delayed the turnover of SRX, (3) Keap1 knockdown impaired the polyubiquitination of SRX, (4) K61 of SRX was the predominant ubiquitin attachment site catalyzed by Keap1. Interestingly, NRF2, as one of the substrates of Keap1, has been recently reported to transactivate SRX40. Our data showed that the downregulation of Keap1 still caused SRX accumulation in some extent after excluding the upregulation of SRX induced by NRF2, indicating that Keap1 specifically regulated the expression of SRX at the protein level (Supplementary Fig. 2a). As a result, our findings not only reveal a mechanism by which SRX is governed by Keap1 at the posttranslational level, but also deliver an in-depth insight into the regulation of SRX in the context of tumorigenesis.
Generally, previous study reported that Keap1 bound and degraded substrates through the Kelch ___domain19,20,22. Surprisingly, in this study, we disclosed that the BACK ___domain of Keap1 was responsible for recruiting SRX, which was distinct from the typical way Keap1 interacted with the substrate. One possible explanation for the difference could be that, SRX, as an antioxidant protein, was bound and ubiquitinated by the BACK ___domain of Keap1, which contains the most reactive residues (C257, C272, C288, and C297) that serve as the direct redox sensors35. Consistently, our data showed that the SRX-binding ability of Keap1 mutant C257Y within the BACK ___domain was remarkably impaired, and the corresponding ubiquitination capacity was significantly weakened. Our findings disclose a mechanism by which Keap1 binds and degrades redox substrates through its noncanonical functional domains.
Keap1, as the best studied E3 ubiquitin ligase, was reported to be tightly associated with tumorigenesis and metastasis by degrading oncogenic substrates41,42, but the mechanisms underlying Keap1 prohibits tumor progression is still poorly understood. In the current study, we identified an oncogenic substrate SRX of Keap1 and manifested a mechanism of Keap1-SRX axis modulating CRC tumorigenesis and metastasis. Our results revealed that downregulation of Keap1 triggered an accumulation of SRX by blocking its ubiquitination, thereby promoting CRC metastasis modulating by the AP-1/MMP9 pathway, which commonly elevated in human cancers43,44. Intriguingly, our research discovered several CRC-derived Keap1 mutants (L223M, C257Y and L291M) exhibiting similar phenotypic features and tumorigenic mechanisms by abrogating SRX degradation. Besides EMT and metastasis, SRX’s interaction with various binding partners allowed it to affect a variety of signaling pathways, such as redox response pathways and EGFR-MAPK cascades45,46. Thus, it might be possible that Keap1 knockdown induced CRC metastasis attributed to the aberrant activation of multiple SRX-engaged oncogenic pathways.
In conclusion, our findings established that Keap1 ubiquitinated and degraded SRX, emphasizing the pivotal role of the Keap1-SRX axis in restraining CRC progression, which underscored the potential of targeting the Keap1-SRX axis as a strategic approach to combat CRC.
Methods
Study approval
Animal experiments were performed in accordance with the National Guidelines for Experimental Animal Welfare, with approval from the Institutional Animal Care and Use Committee of Shanghai Model Organisms Center, Inc. (approval No. 2022-0050). Tumor tissues and adjacent tissues from patients with CRC were obtained from Taizhou Hospital, Taizhou, China. Written informed consent regarding tissue and data use for scientific purposes was obtained from all participating patients. The study was approved by the Research Ethics Committee of Taizhou Hospital (approval No. YB M-05-02) in agreement with the Declaration of Helsinki.
Cell culture
All the cells were obtained from ATCC and maintained at 37 °C in 5% CO2 incubators. HEK293T and SW480 cells were cultured in DMEM medium (Basal Media Technology) supplemented 10% FBS (Aus GeneX). HCT116 cells were cultured in McCoy’s 5A medium (Basal Media Technology) with 10% FBS. HCT15 cells were cultured in RPMI 1640 medium (Basal Media Technology) with 10% FBS. RKO cells were cultured in MEM medium (Basal Media Technology) with 10% FBS.
Plasmids and mutagenesis
To construct Flag-SRX, HA-Keap1 and Myc-Keap1 plasmids, the cDNA of SRX (NM_080725.3) and Keap1 (NM_203500.2) were amplified by PCR and cloned into the vector Flag-pXJ40, HA-pXJ40 and Myc-pXJ40, respectively. Cells were transiently transfected with various plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Keap1 or SRX mutants were generated by Phusion™ Site-Directed Mutagenesis Kit (Thermo Fisher Scientific) and verified by Sanger sequencing. The somatic mutations data for Keap1 and SRX in CRC patients were sourced from the retrieval results on the COSMIC (https://cancer.sanger.ac.uk/cosmic) website.
Antibodies and reagents
Antibodies specific to SRX (Santa Cruz, sc-514940, 1:500; sc-166566, 1:1000; sc-166786, 1:500), Keap1(Cell Signaling Technology, 8047, 1:1000; 4678, 1:1000), NAE1 (Cell Signaling Technology, 14321, 1:1000), p27 (Cell Signaling Technology, 3686, 1:1000), Cullin3 (Cell Signaling Technology, 2759, 1:1000), Cullin4A (Cell Signaling Technology, 2699, 1:1000), p-JNK (Cell Signaling Technology, 9255, 1:1000), p-c-Jun (Cell Signaling Technology, 3270, 1:1000), MMP9 (Cell Signaling Technology, 13667, 1:500; ABclonal, A25299, 1:1000), E-Cadherin (Cell Signaling Technology, 3195, 1:1000; ABclonal, A22333, 1:1000), Snail (Cell Signaling Technology, 3879, 1:1000), Ubiquitin (Cell Signaling Technology, E4I2J, 1:1000; P37, 1:1000), UBA3 (Abcam, ab124728, 1:1000), UBE2M/UBC12 (Abcam, ab109507, 1:1000), ROC1 (Abcam, ab133565, 1:1000), Cullin1 (Abcam, ab75817, 1:1000), Cullin2 (Abcam, ab166917, 1:1000), Cullin4B (Abcam, ab67035, 1:1000), Cullin5 (Abcam, ab184177, 1:1000), NRF2 (Proteintech, 16396-1-AP, 1:1000), Flag-tag (Proteintech, 20543-1-AP, 1:10000), HA-tag (Cell Signaling Technology, 3724, 1:4000), Myc (Cell Signaling Technology, 2276, 1:2000; 2278, 1:2000) β-actin (HuaBio, EM21002, 1:3000), Anti-Flag® M2 Affinity Gel (Merck, A2220), Anti-HA-Tag Mouse antibody (Agarose Conjugated) (Abmart, M20013M) were purchased commercially. MLN4924 (MCE, HY-70062), MG132 (Sigma Aldrich, 474790), cycloheximide (CHX, Cell Signaling Technology, 2112), MMP-9-IN-1 (MCE, HY-135232), and T-5224 (MCE, HY-12270) were dissolved in dimethylsulfoxide (DMSO, MCE, HY-Y0320) and kept at −20 °C.
Label-free quantification
SW480 cells were treated for 12 h with DMSO or MLN4924 (1 μM) with 3 replicates per group, for a total of 6 samples. Trypsin was used to digest lysed cells. Then the peptides were subjected to MS analysis on a Q Exactive system (Thermo Fisher Scientific). Using the MaxQuant software (version 1.6.5.0), raw files were searched against the human SwissProt protein database (20,427 proteins) with FDR < 1% at the peptide and protein level. Variable modifications, such as acetylation (Protein-N terminus) and oxidation methionine (M), and fixed modifications, such as carbamidomethylation (C), both utilized default settings. We quantitated the MS signals using the MaxQuant LFQ algorithm for label-free quantification, and the protein intensities were shown using iBAQ (intensity-based absolute protein quantification)47. After that, the iBAQ of each sample was entered into FOT, which is calculated as the iBAQ of a protein multiplied by the sum of iBAQs for all identified proteins. The normalized abundance of each protein was then represented by the FOT multiplied by 106. The average of 3 repeats was used to quantify the fold change, and the t-test was applied to assess statistical significance.
RNA interference
The cells were transfected with siRNA oligonucleotides (Genepharma) using Lipofectamine RNAi MAX (Invitrogen) according to the manufacturer’s instructions. The siRNA sequences are as follows:
sicontrol: 5′-UUCUCCGAACGUGUCACGU-3′;
siKeap1-1: 5′- GUGGCGAAUGAUCACAGCAAU-3′;
siKeap1-2: 5′- CCUCAAUCGUCUCCUUUAUTT-3′;
siKeap1-3: 5′-GUGCUCAUGUACCAGAUTT-3′;
siNRF2: 5′-GCAUGCUACGUGAUGAAGATT-3′;
siCullin1: 5′-CUAGAUACAAGAUUAUACAUGCGG-3′;
siCullin2: 5′-GCAGAAAGACACACCACAA-3′;
siCullin3-1: 5′- UUGACGUGAACUGACAUCCACAUUC-3′;
siCullin3-2: 5′- UACAUAUGUGUAUACUUUGCGAUCC-3′;
siCullin3-3: 5′- CAACACUUGGCAAGGAGACTT-3′;
siCullin4A: 5′-GAAGAUUAACACGUGCUGG-3′;
siCullin4B: 5′-AAGCCUAAAUUACCAGAAA-3′;
siCullin5: 5′-CUACUGACUCUGAGAAAUA-3′;
siROC1-1: 5′-GACTTTCCCTGCTGTTACCTAATT-3′;
siROC1-2: 5′-CTGTGCCATCTGCAGGAACCACATT-3′;
siNAE1: 5′-GGGUUGUGCUUUAGUCUGU-3′;
siUBA3: 5′-UGUUCUGGUAGCCUGGGCAUAGAUG-3′;
siUBC12: 5′-GGGCUUCUACAAGAGUGGGAAGU-3′;
siSPOP:5′-CAACUAUCAUGCUUCGGAU-3′;
siLZTR1:5′-CGGGACAAGAUGUUUGUAUTT-3′;
siKLHL22: 5′-GGUGUGUCCUACAAUGCUATT-3′.
Cycloheximide (CHX) chase assay
To determine the half-life of SRX, SW480 and HCT116 cells were treated with CHX (50 μg/mL) for the indicated time. SW480 and HCT116 cells were transfected with the indicated siRNAs. Seventy-two hours after transfection, cells were treated with CHX (50 μg/mL), harvested at indicated time points and then subjected to western blotting analysis.
Immunoprecipitation
Before lysis, the cells were treated with MG132 (20 μM) for 6 h. Cells were harvested and lysed in NP-40 lysis buffer (Beyotime Biotechnology) with protease inhibitors (1 mM PMSF; protease inhibitor cocktail, Merck) and protein phosphatase inhibitors (Thermo Fisher Scientific) for 30 min on ice, and centrifuged at 13,400 × g for 15 min. After centrifugation, the supernatant was incubated with anti-SRX antibody overnight at 4 °C. Complexes were pulled down by incubation with protein A/G Agarose Beads (Proteintech) for another 2 h. Cells transfected with Flag-SRX, HA-Keap1, and related mutations were lysed as described above, and then incubated with anti-Flag agarose beads and anti-HA agarose beads overnight at 4 °C, respectively. The immunoprecipitates were washed 3 times with the NP-40 lysis buffer and analyzed by western blotting with the indicated antibodies.
In vivo ubiquitination assay
Cells were treated with MLN4924 or transfected with indicated expression plasmids or indicated siRNA and followed treated with MG132 (20 μM) for 6 h. The collected cells were lysed in a denaturing lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, and 1% SDS with protease and phosphatase inhibitors). Cell lysates were boiled for 10 min, diluted ten times in lysis buffer without SDS, and subjected to immunoprecipitation with anti-SRX antibody or anti-Flag agarose beads followed by western blotting analysis.
Protein purification and GST pull-down assay
The His-SRX plasmid was constructed with the GV296 vector, and the GST, GST-Keap1, and GST-Keap1-BACK plasmids were constructed with the GV205 vector. GV296 and GV205 vectors were purchased from Shanghai Jikai Gene Chemical Technology Co., Ltd., All plasmids were transformed in E. coli BL21 (DE3) strain and cultured in the Luria-Bertani (LB) with Kanamycin. The corresponding fusion protein was induced by 0.5 mM isopropyl-B-D-1-thiogalactopyranoside (IPTG) at 16 °C for 16 h and harvested by GST-tag or His-tag agarose beads (Yeasen Biotechnology) according to the manufacturer’s protocol. Purified His-SRX protein was incubated separately with GST-tag agarose beads, GST, GST-Keap1, and GST-Keap1-BACK in binding buffer (Sangon Biotech) at 4 °C for 4 h, and then immobilized to GST-tag agarose beads at 4 °C overnight. After washing three times with washing buffer (Sangon Biotech) at 500 × g for 1 min, bound proteins were subjected to SDS-PAGE and detected by Coomassie Brilliant Blue staining.
Generation of stable cell lines
The CRISPR/Cas9 system was used to knockdown Keap1 and SRX respectively or simultaneously. The small guide RNA (sgRNA) oligonucleotides specifically against Keap1 and SRX (5 μg) were inserted into vector lenti-guide-puro, respectively, and then were co-transfected with package plasmids psPAX2 (4 μg) and pMD2.G (3 μg) into HEK293T cells by using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, the lentivirus supernatant was collected and filtered, and infected SW480 and HCT116 cells along with 10 μg/mL polybrene (Santa Cruz). Puromycin (2 μg/mL, Invitrogen) was utilized to select the infected cells for 2 weeks. The human sgRNA sequences of Keap1 and SRX are as follows:
sgKeap1-1: 5’-CACCGTCGTAGCCCCCATGAAGCAC-3’;
sgSRX: 5’-CACCGCCACTCGGGCCGCATCGCCG-3’.
Cell viability assay and clonogenic survival assay
Cells were seeded in 96-well plates at a density of 2 × 103 cells per well and cultured for 72 h. Cell viability was determined using the ATP-Lite Luminescence Assay Kit (PerkinElmer) according to the manufacturer’s protocol. For clonogenic survival assay, cells were seeded at 200 cells per well in 6-well plates for 12 days, then the 6-well plates were washed with cold 1×PBS 3 times and fixed with 4% paraformaldehyde at room temperature for 30 min. Colonies on the plates were stained with 0.1% crystal violet for 30 min. Colonies comprising 50 cells or more were counted under an inverted microscope.
Cell migration and invasion assays
For migration assay, cells were suspended in serum-free media and seeded in 24-well plates’ upper chambers per well at a density of 1.5 × 105 cells (HCT116) or 2 × 105 cells (SW480), and then 650 μL of medium supplemented with 20% FBS was added to the lower chambers. For invasion assay, a Matrigel polycarbonate membrane (Corning) was placed in the upper chamber. The migrating cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet solution. Cells on the outside of the upper chambers were captured and counted under a microscope (Thermo Fisher Scientific).
Gelatin zymography
Cells were cultured in serum-free medium and the concentrated culture medium was separated on native SDS-PAGE gel containing 0.1% gelatin. The zymogram was performed following the manufacturer’s instructions (Invitrogen). The enzyme activity was observed as a clear colorless region (band) depleted of gelatin in the gel against the blue background.
Animal model of tumor xenograft
For subcutaneous-transplantation tumor model, 3 × 106 stable HCT116 cells were subcutaneously injected into the right back of 5-week-old female Balb/c nude mice (n = 32, 8 per group). Tumor size was measured with calipers at the indicated time points and calculated as (length × width2)/2. Mice were euthanized at the end of the study, and tumor tissues were harvested, photographed, and weighed. For experimental metastatic model, 2.5 × 106 stable HCT116 cells were injected into 6-week-old female Balb/c nude mice through the tail vein (n = 20, 5 per group). After euthanasia, the lungs were washed with pre-cooled 1×PBS, and then fixed with Bouin’s fixative for 24 h. After fixation, the lung surface nodules were counted and embed the lung tissue for sectioning, followed by H&E staining. For orthotopic mouse model, 7-week-old female NCG mice were anesthetized with 2% isoflurane, and the cecum was exposed through a lower-abdomen incision. A total of 2 × 106 luciferase-expressing stable HCT116 cells were injected subserosally at the cecum wall (n = 20, 5 per group). Eight days after modeling, tumor burden was monitored once every 4 days with an IVIS Lumina III imaging system (PerkinElmer), and mice were euthanized after the fourth imaging (Day 20). Quantifications were made with Living Image v.4.3.1 (PerkinElmer). Mice were kept and bred at a constant room temperature with a 12:12 h light/dark cycle and fed a standard rodent diet and water. The maximal tumor burden permitted by ethics committee was no more than 1500 mm3, and the maximal tumor burden did not exceed the limit.
Tumor tissue collection
For immunohistochemical (IHC) staining assay, fresh primary CRC tissues were collected from 136 CRC patients who underwent resection at Taizhou Hospital (Taizhou, China), including 75 males and 61 females aged between 29 to 86 years old. Among them, 128 CRC patients simultaneously collected their paired adjacent normal tissues, while 8 CRC patients only collected CRC tissues. Ten pairs of CRC tissues and adjacent tissues of patients were lysed for western blotting analysis. For human CRC tissue array analysis, sex/gender was not considered in the research design.
Immunohistochemistry analysis
The human CRC tissue array was subjected to IHC analysis stained with SRX antibody from Shanghai Biochip. The 5-micron-thick tissue array sections were dehydrated and subjected to peroxidase blocking. The primary antibody was added and incubated at room temperature for 30 min using the DAKO AutoStainer and the DakoCytomation EnVision+ System-HRP detection kit (Dakocytomation). After counterstaining with hematoxylin, the slides were observed under a microscope, and images were captured. Immunohistochemistry results was further evaluated by a semiquantitative approach H-score ranging from 0 to 300. For each sample, H-scoring assessment was estimated by multiplying staining intensity (0, negative; 1+, weak; 2+, moderate; and 3+, strong) together with the percentage of positive cells (0–100%) using formula: [1 × (% cells 1+) + 2 × (% cells 2+) + 3 × (% cells 3+)]. Based on the staining intensity, we classified the samples into four groups: the weakest group (+), weak group (++), medium group (+++), and strongest group (++++). Interpretation of IHC results was performed by two independent pathologists.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 9 software (GraphPad Software). Data were presented as the mean ± SD. An unpaired two-tailed Student’s t test was used to compare parameters between groups. Survival was analyzed using the Kaplan–Meier method, and data were compared using the log-rank test with Statistical Program for Social Sciences (SPSS) software version 25.0.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available in the article and its Supplementary Information files. The mass spectrometry proteomics data generated in this study have been deposited in ProteomeXchange Consortium via the iProX partner repository under accession code PXD048951. Source data are provided with this paper.
References
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 72, 7–33 (2022).
Wang, Y.-N. et al. CPT1A-mediated fatty acid oxidation promotes colorectal cancer cell metastasis by inhibiting anoikis. Oncogene 37, 6025–6040 (2018).
Cook, A. D., Single, R. & McCahill, L. E. Surgical resection of primary tumors in patients who present with stage IV colorectal cancer: an analysis of surveillance, epidemiology, and end results data, 1988 to 2000. Ann. Surg. Oncol. 12, 637–645 (2005).
Schmoll, H. J. et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann. Oncol. 23, 2479–2516 (2012).
Böckelman, C., Engelmann, B. E., Kaprio, T., Hansen, T. F. & Glimelius, B. Risk of recurrence in patients with colon cancer stage II and III: a systematic review and meta-analysis of recent literature. Acta Oncol. 54, 5–16 (2015).
Holch, J. W. et al. Pattern and dynamics of distant metastases in metastatic colorectal cancer. Visc. Med. 33, 70–75 (2017).
Aparicio, C. et al. Cell therapy for colorectal cancer: the promise of chimeric antigen receptor (CAR)-T cells. Int. J. Mol. Sci. 22, 11781 (2021).
Sun, Y., Hegamyer, G. & Colburn, N. H. Molecular cloning of five messenger RNAs differentially expressed in preneoplastic or neoplastic JB6 mouse epidermal cells: one is homologous to human tissue inhibitor of metalloproteinases-3. Cancer Res. 54, 1139–1144 (1994).
Rhee, S. G., Jeong, W., Chang, T. S. & Woo, H. A. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int. Suppl. 106, S3–S8 (2007).
Ogata, F. T. et al. Thiol-based antioxidants and the epithelial/mesenchymal transition in cancer. Antioxid. Redox Signal. 36, 1037–1050 (2022).
Jiang, H. et al. Sulfiredoxin promotes cancer cell invasion through regulation of the miR143-Fascin axis. Mol. Cell Biol. 42, e0005122 (2022).
Ni, D., Zhou, H., Wang, P., Xu, F. & Li, C. Visualizing macrophage phenotypes and polarization in diseases: from biomarkers to molecular probes. Phenomics 3, 613–638 (2023).
Wei, Q. et al. Sulfiredoxin-Peroxiredoxin IV axis promotes human lung cancer progression through modulation of specific phosphokinase signaling. Proc. Natl Acad. Sci. USA 108, 7004–7009 (2011).
Lan, K. et al. Sulfiredoxin may promote cervical cancer metastasis via Wnt/β-catenin signaling pathway. Int. J. Mol. Sci. 18, 917 (2017).
Ramesh, A., Varghese, S. S., Doraiswamy, J. & Malaiappan, S. Role of sulfiredoxin in systemic diseases influenced by oxidative stress. Redox Biol. 2, 1023–1028 (2014).
Kobayashi, A. et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 24, 7130–7139 (2004).
Koppula, P. et al. A targetable CoQ-FSP1 axis drives ferroptosis- and radiation-resistance in KEAP1 inactive lung cancers. Nat. Commun. 13, 2206 (2022).
Dinkova-Kostova, A. T., Kostov, R. V. & Canning, P. Keap1, the cysteine-based mammalian intracellular sensor for electrophiles and oxidants. Arch. Biochem. Biophys. 617, 84–93 (2017).
Chen, Y. et al. Disruption of the Keap1-mTORC2 axis by cancer-derived Keap1/mLST8 mutations leads to oncogenic mTORC2-AKT activation. Redox Biol. 67, 102872 (2023).
Siswanto, F. M., Oguro, A. & Imaoka, S. Sp1 is a substrate of Keap1 and regulates the activity of CRL4AWDR23 ubiquitin ligase toward Nrf2. J. Biol. Chem. 296, 100704 (2021).
Lee, D.-F. et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol. Cell 36, 131–140 (2009).
Lo, S.-C. & Hannink, M. PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. J. Biol. Chem. 281, 37893–37903 (2006).
Zhou, Y. et al. FAM117B promotes gastric cancer growth and drug resistance by targeting the KEAP1/NRF2 signaling pathway. J. Clin. Investig. 133, e158705 (2023).
Xu, K. et al. Battles against aberrant KEAP1-NRF2 signaling in lung cancer: intertwined metabolic and immune networks. Theranostics 13, 704–723 (2023).
Zhu, L. et al. Chaperone-mediated autophagy degrades Keap1 and promotes Nrf2-mediated antioxidative response. Aging Cell 21, e13616 (2022).
Tian, H. et al. Keap1: one stone kills three birds Nrf2, IKKβ and Bcl-2/Bcl-xL. Cancer Lett. 325, 26–34 (2012).
Nakayama, K. I. & Nakayama, K. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 (2006).
Deshaies, R. J. & Joazeiro, C. A. P. RING ___domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009).
Chen, Y. et al. Phosphorylation regulates cullin-based ubiquitination in tumorigenesis. Acta Pharm. Sin. B 11, 309–321 (2021).
Wu, K. et al. Inhibitors of cullin-RING E3 ubiquitin ligase 4 with antitumor potential. Proc. Natl Acad. Sci. USA 118, e2007328118 (2021).
Enchev, R. I., Schulman, B. A. & Peter, M. Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 16, 30–44 (2015).
Zhou, L., Zhang, W., Sun, Y. & Jia, L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell. Signal. 44, 92–102 (2018).
Toth, J. I., Yang, L., Dahl, R. & Petroski, M. D. A gatekeeper residue for NEDD8-activating enzyme inhibition by MLN4924. Cell Rep. 1, 309–316 (2012).
Cheng, J. et al. Functional analysis of Cullin 3 E3 ligases in tumorigenesis. Biochim. Biophys. Acta Rev. Cancer 1869, 11–28 (2018).
Kopacz, A., Kloska, D., Forman, H. J., Jozkowicz, A. & Grochot-Przeczek, A. Beyond repression of Nrf2: an update on Keap1. Free Radic. Biol. Med. 157, 63–74 (2020).
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48 (2023).
Hao, Y. et al. Critical role of the sulfiredoxin-peroxiredoxin IV axis in urethane-induced non-small cell lung cancer. Antioxidants 12, 367 (2023).
Barquilha, C. N. et al. Sulfiredoxin as a potential therapeutic target for advanced and metastatic prostate cancer. Oxid. Med. Cell Longev. 2020, 2148562 (2020).
Soriano, F. X. et al. Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Mol. Cells 27, 279–282 (2009).
Mishra, M. et al. Nrf2-activated expression of sulfiredoxin contributes to urethane-induced lung tumorigenesis. Cancer Lett. 432, 216–226 (2018).
Lignitto, L. et al. Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell 178, 316–329 (2019).
Jeong, Y. et al. Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance. Cancer Discov. 7, 86–101 (2017).
Liu, J.-F., Chen, P.-C., Chang, T.-M. & Hou, C.-H. Monocyte chemoattractant protein-1 promotes cancer cell migration via c-Raf/MAPK/AP-1 pathway and MMP-9 production in osteosarcoma. J. Exp. Clin. Cancer Res. 39, 254 (2020).
Li, S. et al. Cholic acid stimulates MMP-9 in human colon cancer cells via activation of MAPK, AP-1, and NF-κB activity. Int. J. Mol. Sci. 21, 3420 (2020).
Jiang, H. et al. Sulfiredoxin promotes colorectal cancer cell invasion and metastasis through a novel mechanism of enhancing EGFR signaling. Mol. Cancer Res. 13, 1554–1566 (2015).
Mishra, M., Jiang, H., Wu, L., Chawsheen, H. A. & Wei, Q. The sulfiredoxin-peroxiredoxin (Srx-Prx) axis in cell signal transduction and cancer development. Cancer Lett. 366, 150–159 (2015).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Acknowledgements
This work was supported by the National Key R&D Program of China (2022YFC3500200 to L.J., 2022YFC3500202 to L.J. and 2023YFC3606600 to S.W.), National Natural Science Foundation of China (82372984 to L.J., 82272987 to S.W. and 82172933 to S.W.), Shanghai Frontiers Science Center of Disease and Syndrome Biology of Inflammatory Cancer Transformation (2021KJ03-12 to L.J.), Shanghai Clinical Research Center of Traditional Chinese Medicine Oncology, Science and Technology Commission of Shanghai Municipality (21MC1930500 to L.J.), Science and Technology Commission of Shanghai Municipality Rising-Star Program (23QA1403000 to S.W.), and Shanghai Municipal Health Commission Excellent Program (20244Z0018 to S.W.). We thank Ping Wang and Lan Fang from Tongji University Cancer Center for providing us with the His-ub and HA-ub plasmids.
Author information
Authors and Affiliations
Contributions
Conceptualization: S.W. and L.J.; methodology: F.Z., L.S.L. and Y.P. (in vitro); F.Z. and Y.C. (in vivo); L.H.L. and L.C. (Label-free quantification); H.Z. (Clinical sample collection); X.Z. and W.Z. (IHC analysis); S.W. and F.Z. (statistical analysis); writing: F.Z. and S.W.; supervision: S.W. and L.J. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Masahiro Aoki, Qiou Wei and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, F., Li, L., Chen, Y. et al. CRL3Keap1 E3 ligase facilitates ubiquitin-mediated degradation of oncogenic SRX to suppress colorectal cancer progression. Nat Commun 15, 10536 (2024). https://doi.org/10.1038/s41467-024-54919-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54919-2