Abstract
Microorganisms are present in snow/ice of the Antarctic Plateau, but their biogeography and metabolic state under extreme local conditions are poorly understood. Here, we show the diversity and distribution of microorganisms in air (1.5 m height) and snow/ice down to 4 m depth at three distant latitudes along a 2578 km transect on the East Antarctic Plateau on board an environmentally friendly, mobile platform. Results demonstrate the widespread distribution of microorganisms in the ice down to at least 4 m depth. Data point to geochemical and bacterial geographic distribution that correlate with wind trajectory and speed, modulated by local gathering and recirculation of microorganisms through snow drifting. Reservoir effects and community selection appear to occur over time, favoring microorganisms best adapted to hypothermal and hyperarid conditions. A new cyanobacterial species (Gloeocapsopsis sp) was isolated from 3 to 4 m depth. Our findings suggest that some microorganisms could exhibit transient, basal metabolic activity when associated to high salt particles, contributing to set biodiversity patterns and biogeographic compartmentalization on Antarctic Plateau ice.
Similar content being viewed by others
Introduction
The Antarctic Plateau is one of the most extreme environments on Earth. It has recorded the lowest temperature on Earth at Vostok station (-89.2 °C, although it is estimated that it can reach -93.2 °C) and is one of the sites with the lowest precipitation record1,2. The low temperature and absolute humidity, together with the high UV radiation during summer, scarcity of liquid water and oligotrophy, make the Antarctic Plateau an excellent natural laboratory to investigate the extremes of life on Earth. Near the South Pole, the water-vapor content of the air is extremely low, with mean annual atmospheric values ranging between 200 and 700 precipitable micrometers (pr µm)3,4, approaching Mars-like conditions5.
Snow/ice in the Antarctic Plateau offers an archive of past climatic and atmospheric events6,7 as well as historical accumulation of biological material. Few studies have focused on the microbiology of the high Antarctic Plateau, and those that have done so have only explored surface snow/ice8 down to 30 cm deep9 or in subglacial lakes10. Several reports show that microbial communities are heterogeneously distributed in surface snow at different locations next to scientific stations, all with a near coastal influence in Eastern9,11,12 and Western Antarctica13. Previous studies report the presence of 102-103 cells ml-1 in surface snow at South Pole Station, presenting tantalizing evidence that microorganisms could have a basal metabolism in snow/ice at -12 to -17 °C14,15. However, the possibility of in situ metabolic activity in this region has been questioned on the grounds that the snow/ice would not contain enough liquid water under local conditions15. Thus, microbial diversity and ecology in snow/ice layer of the interior of the Antarctic Plateau remain understudied, and the metabolic state of those microorganisms is unknown. Aerial dispersion has been considered the main input of microbial particles, and atmospheric transport is claimed to be the origin of community heterogeneity, which partially explains microbial biogeography. Antarctic Plateau snowpack is a big genetic reservoir of potentially active microorganisms9,11 although there is no conclusive evidence supporting this hypothesis.
Some studies report that successful global distribution of microorganisms, including of Antarctica, is a consequence of wind circulation and air dispersion16, while others suggest constrained distribution to particular regions due to the predominance of local climatic and geographic factors17. We hypothesize that microbial biogeography in the Antarctic Plateau, despite its apparent uniformity, is highly influenced by regional factors such as wind circulation, aerosol deposition17,18,19, and the resistance of life under such extreme conditions during the airborne traverse and residence in the ice. To test this hypothesis, we conducted a large-scale survey of geochemistry and microbiology from the air (ground level) to 4 m depth in the snow/ice layer along a 2578 km transect on the East Antarctic Plateau.
In the following Results sections we describe: (i) how we resolve the logistical constraints of accessing, sampling and doing in situ experiments on the Antarctic Plateau by using a unique, wind-powered mobile science station called WindSled; (ii) the measurements of environmental and geochemical parameters that are critical to understand local and regional biodiversity, as well as experiments carried out during the cruise aimed at assessing the possibility of deliquescence events in the presence of salts and their potential effects on life; (iii) on site detection of microorganisms using a portable immunosensor (LDChip, Life Detector Chip), designed for rapid in field life detection and for planetary exploration20; and (iv) the bacterial diversity and its correlation with wind patterns to infer any geographical distribution along the transect, including enrichment cultures of viable bacteria from snow/ice samples.
Results
Exploring the Antarctic Plateau with an environmentally friendly mobile scientific station
Over the 2018-2019 austral summer, we carried out a 2578 km scientific exploration campaign into the Northeastern Antarctic Plateau, between Novolazarevskaya Station (120 m a.s.l.) and Dome Fuji ( > 3,500 m a.s.l.)21, using the WindSled mobile platform (Fig. 1A, B). Crewed by four well-trained people, WindSled moved 200 kg of scientific instrumentation to perform multiple sampling and in field scientific experiments. Air temperature ranged from -14.2 °C to -42.2 °C along the transect. Six snow/ice cores were obtained down to 4 m depth along the transect for isotopic analyses (see Methods), and three sets of samples named N (North), M (Middle), and S (South) were obtained for detailed geochemical and microbiological studies. The latter set included: (i) airborne material at 1.5 m height and, (ii) snow/ice cores from 0 to 4 m depth (Fig. 1 A–D, Table 1).
A Itinerary showing the locations where the two set of samples were collected: red circles named N, M and S, for three cores for geochemical and microbiological studies; and yellow stars (a–f) for six oxygen and hydrogen isotope analysis core samples (see Fi. 2). B the WindSled mobile platform consisting of three modules (driving, payload, and living room) and wind-powered by a 200 m2 kite. C Drilling system and the aspect of the 1 m snow/ice cores. D Airborne aerosol collector in the field and scheme: 1, manifold pipe; 2, turbine; 3; sample holder; 4, Fan 9GT0612P4G001; 5, collector support; arrow, flow direction (see Materials and Methods for further details). E Five-day back-trajectories of the air parcels from the collected samples on the African sector of the East Antarctic Plateau. Grey contour lines indicate altitude at 500 m intervals. The depicted back-trajectories originate from the three distinct sampling points: N (red), M (green) and S (blue). These trajectories are computed at 25%, 50%, and 75% of the boundary-layer height with hourly initialization the sampling periods.
Reconstructed five-day back-trajectories of air masses along the entire traverse region are shown in Fig. 1E. PERMANOVA and Bonferroni multiple comparison tests showed significant evidence of heterogeneity among the three sets of back-trajectories (all p-values < 0.0001). Air trajectories at the N and M sites crossed the region of the plateau near the coast of Queen Maud Land in the hours before sampling. Air masses reaching sampling site M showed a flow origin nearby the Amery Ice Shelf. The set of trajectories was parallel to the coast at the edge of the Plateau. On the other hand, the southernmost sampling site S was dominated by internal continental airflows circulating around Dome Fuji, with minimal coastal contribution. To rule out exceptional or anomalous meteorological events, we analyzed the degree of circulation analogy of each sampling day (Supplementary Fig. 1). Results showed that each sampling day contained a significant number of analogs, amounting to more than 6% of total. This suggests that the meteorological conditions influencing the back-trajectories observed in the three sampling areas were typical weather patterns in the region during the austral summer.
The δ18O of water recovered from melted ice/snow core samples showed a characteristic 18O depletion towards higher altitudes, which is consistent with a fractionation against the heaviest O isotope as wind masses traverse the continent (Fig. 2A, B and Supplementary Table 1). Snow/ice core samples contained low molecular weight organic acids (acetate and propionate) and inorganic anions (chloride, sulfate, nitrate) at low concentration (ppm) (Fig. 2C), with peaks of chloride at 2-3 m depth in all sampling sites, but specially at M (4.7 ppm), sulfate (0.85 ppm), and nitrate (2.5 ppm). Sample M also showed relatively high concentration of Na at 2-3 and 3-4 m depth, coincident with the peaks of chloride and sulfate. These results, together with the detection of Mg, Ca, and K (Supplementary Table 2), indicated that the M site could have a higher costal influence than N and S sites.
A δ18O in water from melted snow/ice cores from north to south collected along the transect (a-f). Ratio δ18O is reported as per mil relative to Standard Mean Ocean Water (SMOW). The plot shows the δ18O values obtained from core samples every ten centimeters intervals from the drills performed at 6 locations (a-f) at the indicated coordinates. Each sample series is represented in a different color, from light grey for the northernmost sample to deep red for the southernmost one. The plot shows a 18O depletion as a function of the distance from the coast (N, north) to the interior of the Antarctic Plateau (to the South and East). B Mean δ18O values at each site e-f (diamonds with color gradient as in A) as a function of latitude, with N, M, and S sites located over the curve (black dots) as a guidance. Error bars correspond to standard deviation (SD) of all measurements at different depths in a single site (site a, n = 30; site b, n = 40; site c, n = 35; sites d and e, n = 40 each one, and; site f, n = 37). See Supplementary Table 1). C Ion chromatography analysis of melted and filtered water samples showing inorganic and short-chain organic anions at the three sites: North (red) DN1-4, Middle (green) DM1-4, and South (blue) DS1-4, from 0-1 m (1), 1-2 m (2), 2-3 m (3), and 3-4 m (4) core depths. Bars show mean values of at least two (n = 2) technical measurements (overlayed points) at different dilutions from each 1-m melted core sample.
Experiments during the cruise revealed NaCl-associated deliquescence at sub-freezing temperatures
We measured temperature and RH relative to ice inside pure NaCl (TNaCl and RHice_NaCl, respectively) exposed to conditions on the Antarctic Plateau to determine whether deliquescent brines could form at any point along the transect (Fig. 3A, B). RHice_air values oscillated from a maximum of 93% to a minimum of 37% (transect average = 81.8%), likely due to ice sublimation (Fig. 3B). Within the salt, TNaCl oscillated from a maximum of +4 °C to a minimum of –42 °C (transect average = –22 °C), whereas RHice_NaCl oscillated from a maximum of 81% to a minimum of 12% (transect average = 56%). Three periods were recorded when RHice_NaCl remained constant between 75% and 80%, the deliquescence point of NaCl, for several days. During these periods, TNaCl changed following daily oscillations but reached daily maximums above the eutectic temperature of saturated NaCl brines. The strongest episode lasted approximately three days (between 01/20/2019 and 01/23/2019) and the temperature within the salt oscillated between –17 °C and –39 °C (average temperature = –29 °C). Hence, during these brief periods environmental conditions were compatible with the formation of NaCl deliquescent brines. We note that these observations represent a rough approximation to conditions in the air near the snow/ice surface.
A Deliquescence experiment. A plastic container filled with NaCl and sensors for conductivity (C), relative humidity (RH) and temperature (T) inserted in the salt. B Values of RHice and T measured in the air (dotted lines) and within NaCl (solid lines) during the transect across the Antarctic Plateau from 12/21/2018 to 02/01/2019. Black areas show periods when RHice within the salt remained constant, and above the equilibrium RH of saturated NaCl brines. This could be due to the formation of thin briny films within the salt via deliquescence. During those periods, the temperature within the salt transiently rose above the eutectic (grey areas). Also indicated are the deliquescence RH and eutectic temperature of saturated NaCl brines. C On site fluorescence sandwich immunoassay (FSIA) with LDChip at M site, showing a portion of LDChip with several positive immunodetections on triplicate spot pattern. Cartoon shows a sandwich immunoassay with an immobilized antibody (Ab), the target biomarker captured (yellow figure), and the fluorescent tracer antibody (FTAb). Squares and numbers indicate the antibodies whose relative fluorescence units where plotted with the entire LDChip results after a single assay with each of 1 m section of M drill (0-1 m, 1-2 m, 2-3 m, 3-4 m) on the right (D), which correspond to: 1, Acidithiobacillus thiooxidans; 2, Tessaracoccus lapidicaptus (Actinobacteriota); 3, Bacillus subtilis spores (Firmicutes); 4, Bacillus subtilis biofilm (Firmicutes); 5, Leptolyngbya boryana (Cyanobacteria); 6, GroEL chaperonine; 7, NifD nitrogenase component; 8, DhnA drought, low temperature, high salinity protectant dehydrin from cyanobacteria; 9, Acidithiobacillus ferrooxidans; 10, NirS nitrite reductase protein; 11, environmental biofilm enriched in iron oxidizing bacteria; 12, Arcobacter sp.
Onboard LDChip immunoassay detected microbial markers in snow/ice cores
Microbial biomass is, together with sea-salt, an important component of Antarctic aerosols that can accumulate on the snow/ice ground at heterogeneous patterns. Melted snow/ice core samples from the M site were tested with the LDChip multiplex immunoassay system (see Methods) to search for microbial markers. The data were analyzed in near real time by a remote science team, which reported positive immunodetections with antibodies raised against microbial strains from Actinobacteriota (Tessaracoccus spp.), Firmicutes (Bacillus spp.), Cyanobacteria (Leptolyngbya spp.), and Proteobacteria. In addition, LDChip detected microbial biopolymeric material and proteins, with the highest intensity in the 2-3 m depth sample (Fig. 3C, D). Additionally, the entire set of samples (air and snow/ice cores) were later analyzed with the LDChip in the laboratory. The results confirmed those obtained in situ and revealed additional positive immunodetections of microbial markers from different phylogenetic groups (Supplementary Fig. 2; Supplementary Table 3).
Bacterial diversity and geographical distribution by DNA sequencing
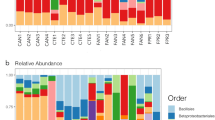
Massive 16S rRNA gene sequencing of air and snow/ice core samples showed a diverse bacterial community, including taxa already detected with the LDChip. We obtained 935,040 total bacterial sequences after quality reads filtering and removal of non-bacterial entities, singletons, and sequences considered as contaminants (See methods). Sequence analysis resulted in 1864 different bacterial ASVs, of which 61 were from air samples, 1799 from snow/ice cores, and 14 were shared between the air and cores. Rarefaction curves showed a good sample coverage (Supplementary Fig. 3). In air samples, Proteobacteria dominated at the N and M sites (67 ± 8%), while both Proteobacteria (41%) and Actinobacteriota (41%) co-dominated at the S site (Fig. 4A). The most abundant genus in AS was Hymenobacter, in AM, Phyllobacterium, and in AS, Brevibacterium (Supplementary Data 1). In contrast to air samples, Firmicutes and Actinobacteriota showed the highest relative abundances in most of the snow/ice cores (46 ± 11% and 27 ± 12%, respectively) except at the N site at 4 m depth, where Proteobacteria dominated (62%), also in agreement with LDChip results (Supplementary Fig. 2). Bacteroidota were detected in all snow/ice cores (4.5 ± 2.9%) and in the air at N site (15%). Consistent with LDChip results, Cyanobacteria were detected in snow/ice cores (1.1 ± 0.5%), at a significantly higher relative abundance at 4 m depth at the S site (DS4 sample, 16%), with Chroococcidiopsis as the most abundant cyanobacterial genus. The most abundant bacterial genera in DN were Corynebacterium (in DN1-3) and Pantoea (in DN4); in DM, Brachybacterium (in DM1-2) and Corynebacterium (in DM3-4); and in DS, Bacillus (in DS1), Staphylococcus (in DS2 and DS4) and Corynebacterium (in DS3) (Supplementary Data 1).
A Bacterial diversity profile after 16S rRNA gene sequence analysis at the phylum level in the three air (AN, AM, AS) and three snow/ice cores (DN, DM, DS) from 1, 0-1 m; 2, 1-2 m; 3, 2-3; 4, 3-4 m depth. B Spatial distribution and relative abundances (%) of bacterial ASVs shared between the air and snow/ice core samples. Relative abundances of ASVs were calculated over the total bacterial community in each sample. (P) Proteobacteria, (F) Firmicutes and (A) Actinobacteriota. Sample code ID is as in A. C Non-metric multidimensional scaling (NMDS) ordination based on Bray-Curtis dissimilarity distances of ASV abundances from air (triangles) and snow/ice (dots) samples from North (red), Middle (green) and South (blue) sampling sites (see text for explanation).
The cell number in the air, as estimated by flow cytometry from collected samples, was 171.62 cells/m3 and 66.65 cells/m3 in N and M sites, respectively, while fell below the detection limit in S. More than 62% of the counted cells showed membrane integrity, indicating a similar percentage of cell viability. Bacterial diversity in air samples was lower (H’ values from 1 to 2.8) than those in snow/ice (H’ values from 3.9 to 5.2) which, in turn, continued to increase with depth down to 3 m (Supplementary Fig. 4). Similarly, richness was higher in the snow/ice cores (from 84 to 359 ASVs) than in the air samples (from 9 to 33 ASVs). Air and snow/ice cores shared 14 ASVs, which represented 23% and 0.8% of the total air and snow/ice core sequences, respectively (Fig. 4B and Supplementary Results for details).
Non-metric multidimensional scaling (NMDS) separated air bacterial communities from the well-defined group of snow/ice communities (Fig. 4C). Snow/ice samples from all three sites clustered together, with a clear sub-group within the northernmost N and M samples. Air samples were farther apart from each other and distant from the snow/ice cores, with AS falling closer to snow/ice samples.
Viable cyanobacteria at 4 m depth in the Antarctic Plateau
The detection of cyanobacterial biomarkers with LDChip and DNA sequences led us to test the potential viability of cells in the filtered samples. Pieces from the 0.22 µm nylon filter from sample DS4, which yielded the highest number of cyanobacterial sequences, were incubated with BG11 medium (see Methods). After 15 days of incubation, a visible green color was observed at the bottom of the flasks, with a growth pattern similar to that described in microbialites. Three-D growth patterns under bright field (Fig. 5A) and fluorescence microscopy (Fig. 5B), together with maximum likelihood phylogenetic analyses (Fig. 5C), suggested that the isolated cyanobacterial strain is a new species of the genus Gloeocapsopsis (Chroococcales, Chroococcaceae) (Genebank No. OR504179), validating the immunological results obtained with LDChip in the field.
A Differential Interference Contrast micrograph showing Gloeocapsopsis sp. CAB1 cells, rarely solitary, mostly arranged in irregular colonies, composed of densely, irregularly aggregated cells, surrounded by mucilaginous envelopes. Cells sub spherical, irregular rounded in outline. Cell duets or tetrads present but are rare. Cell division is irregular in various planes in successive generations, forming round structures with twelve or more cells. B Phycobiliproteins and chlorophyll autofluorescence of the same group of cells shown in A, unveiling differences in chlorophyll content and/or photosynthetic activity. Note also the concentric thylakoids (brightest spots) in some of the cells. C Maximum Likelihood phylogenetic tree from the complete 16S rRNA gene sequence that allowed to classify this strain as a new species within the genus Gloeocapsopsis.
Discussion
Exploring microbial biogeography in snow/ice on the East Antarctic Plateau on board an environmentally friendly mobile scientific station
The environmentally friendly WindSled, is a unique mobile platform that allows to conduct scientific research on the Antarctic Plateau within the environmental framework specified in the Antarctic Treaty22. The high versatility and mobility of WindSled enabled microbial biogeography study over thousand km transects as the one reported herein. On one side, the air samples provided the airborne microbes reaching the plateau at the moment of sampling. On the other, snow/ice cores down to 4 m depth provided a biological record spanning 10 s of years, as inferred from previous estimations of annual snow accumulation rates of 5 to 20 cm y-123,24,25.
Although the lack of samples replicates makes it difficult to associate variation between samples and sites, the NMDS analyses based on Bray-Curtis dissimilarities of Amplicon Sequencing Variants (ASVs) clearly separate the airborne communities between themselves, and from the snow/ice samples, which tended to cluster together (Fig. 4C). These results suggest that significantly different air masses originating in different regions of the Antarctic Plateau transport distinct ensembles of microorganisms. Based on reconstructed air-trajectories 5 days prior to sampling, air samples collected at the N and M sites were potentially exposed to coastal and ice-free regions, which could enrich air with aerosols, including cells, of terrestrial and oceanic origin. In addition, air masses affecting both sites partially converged on their route. On the other hand, wind patterns at the southernmost S site displayed trajectories far from the coast swirling round the dome (Fig. 1E). Such trajectories suggest local recruiting and recirculation of microorganisms through the main physical processes occurring on the Antarctic Plateau, snow drifting and blowing snow. The wind circulation patterns would explain why airborne communities at the N and M sites were more similar and distinct from those at the S site (Fig. 4C). The fact that we could not count visible, intact cells by flow cytometry in the S air sample (AS) indicated that the cells could be embedded in compact aerosol particles difficult to stain and visualize.
Despite the low richness, the different number of bacterial ASVs shared with snow/ice core samples (Fig. 4B) in AN versus AM indicated certain variability in airborne communities between N and M sites. Such variability could be attributed to differences in air velocity, higher at the N site. Faster winds travel longer distances during a given period, which allows to incorporate more diverse particles, including microorganisms, from more distant regions before dry deposition occurs26. Finally, coastal effects and higher katabatics, which promote sublimation changes, also explain the wide variations in the δ18O values with depth at northernmost N site (Fig. 2A). Periods with predominant coastal winds would supply water enriched in heavy oxygen (higher δ18O), aerosols, sea water material27, and consequently bioburden.
Unlike air samples, snow/ice microbial communities were similar at the different sampling locations (N, M, and S) and with depth (1-4 m), so they all clustered together in the NMDS plot (Fig. 3C). Similar vertical bacterial community profiles in snow/ice samples point to a reservoir effect of bacterial cells in the snow/ice layers over time, coming from a mix of air mass trajectories. This suggests a certain degree of community selection in snow/ice over time, preferentially favouring microorganisms that are better adapted to hypothermal and hyperarid conditions such as Firmicutes, Actinobacteria, and Proteobacteria, which are commonly found in desert soils. Hence, the top snow/ice layer in the Antarctic Plateau appears to work as a sedimentation trap of airborne microorganisms, accumulating, selecting, and preserving them over time. This bacterial reservoir capacity of the snow/ice may explain the higher richness and diversity compared to the very low cell concentration and richness in the Plateau air samples (see above and Supplementary Fig. 4). Altogether, the separation of air sequences between them and the slight clustering of the northernmost (N) drill from the other snow/ice samples indicates that there is a certain fractionation of airborne microbiology and, consequently, a differential geographic distribution. This agrees with other work reporting that in the Dry Valleys region airborne connectivity is very limited, allowing certain compartmentalization and the creation of unique, relatively isolated, ecosystems17.
Selection and fractionation processes in the snow/ice are supported by the presence of cyanobacteria down to 4 m depth, with particularly abundant cyanobacterial DNA sequences in DS4 (16%) compared to other samples. Cyanobacteria are found in diverse ice-based ecosystems in Antarctica28, such as ice-shelves29,30 or cryoconites in the surface of glaciers31, where local ice melting helps to maintain a bacterial metabolic minimum. We isolated and cultured a new cyanobacterium of the genus Gloeocapsopsis (strain CAB1) from the DS4 sample, a genus already described in ice-free coastal regions where they colonize soils, stones, and rocks32. Cyanobacteria can accumulate compatible solutes such as sucrose and trehalose or extracellular polymeric substances (EPS) to lower the freezing point of water33,34,35. Moreover, dark pigments in aerosol particles could contribute to a local decrease in albedo, acquiring some radiation that, together with deliquescence, could contribute to some biological activity in the Plateau. Compatible solutes, EPS and strong pigmentation may help cyanobacterial clusters from Gloeocapsopsis or Chroococcidiopsis genera withstand freezing and desiccation during atmospheric transport across the Plateau and remain viable in the snow/ice for relatively long periods of time (10s-100s years). Similarly, microalgae and other microorganisms are part of the aerosol particulate content and many of them are well adapted to low temperatures by producing anti-freezing compounds (e.g., membrane lipids, polyalcohols, exopolysaccharides, compatible solutes) that lower the water freezing point and even could promote deliquescent events at their surface together with salt crystals at microniches36,37. Microbial communities can be trapped in the ice for decades and be capable of obtaining enough water from brine pockets created in a matrix of EPS and salt36, that could give them a survival advantage.
It is expected that the freezing temperatures in the Antarctic Plateau (annual average in Dome Fuji is -52.3 °C) preclude any metabolic activity in the ice-trapped bacterial communities. However, we cannot rule out a minimal bacterial metabolism at a microscale in meltwater veins around salt crystals or other impurities carried by the wind from coastal regions. This instance is supported by deliquescence experiment on WindSled (Fig. 3A, B), and other studies38. The relatively high chloride concentration between 2 and 3 m depth in all samples (Fig. 2C), particularly in sample DM3 (4.7 ppm), is consistent with the occasional deposition of sea salt aerosols, as has been reported by others18. Whether the volume of deliquescent brine is sufficient for bacterial metabolism cannot be conclusively established from our data.
The extreme conditions in the Antarctic Plateau along with the interaction of snow/ice and the role of salts on the feasibility of liquid brines make the Antarctic Plateau one of the best terrestrial analogues to study the habitability of the large icy areas on Mars39,40. The discovery of relatively high concentrations of deliquescent salts (e.g. NaCl, MgClO4, CaClO4)41 in the Martian regolith has sparked renewed interest in the possibility that thin films of low eutectic brines might form episodically in near-surface regolith or ice during warm days when air absolute humidity and ground temperature approach the stability boundary of saturated salt solutions42,43,44. The types of temperature- and humidity-driven phase transitions can be studied in the Antarctic Plateau under dynamic hypothermal and hyperarid conditions (Fig. 3A, B).
The WindSled crew was trained to perform multiplex microarray immunoassays with the LDChip, a 200-antibody microarray test which is the core sensor of the Signs Of Life Detector (SOLID) instrument developed for life detection in planetary exploration45,46. We demonstrated once more the capability of the LDChip for detecting microbial markers on site and the corroboration of these results in the laboratory, what reinforces its utility as a payload instrument for life detection missions to Mars47 or Jupiter’s moon Europa48. Moreover, LDChip can be a valuable tool for immunological assay to monitor human health risks and biohazards in manned missions.
Methods
The WindSled
The WindSled is a unique mobile vehicle created by the Spanish polar explorer Ramón Hernando de Larramendi during 16 years of development. Its basic structure consists of several wooden platforms with crossbars and rails (following the model of the classic Inuit sleds), kites of different sizes, and tents designed as living and working space (e.g., a laboratory) (Fig. 1B). The platform displacement is powered by wind and photovoltaic panels that feed the equipment (See Supplementary Material and Methods for details).
Snow/Ice and Air Sampling
Two sets of 1 m-long and 45 mm in diameter drill cores were sampled sequentially from the surface down to 4 m depth at each site (Fig. 1C). Set one, for oxygen and hydrogen stable isotope determination, consisted of taking snow/ice cores down to 4 m depth at 6 locations (a-f in Fig. 1A) along the transect, fractionated every 10 cm, melted, filtered by 0.22 microns, and stored in 5 mL falcon tubes at ambient (freezing) temperature. Set two, for geochemical and microbiological studies, was collected only at three specific localities: North (N), coordinates 73.17500000 S, 11.55444444 E; Mid-latitude (M), coordinates 77.00777778 S, 18.64750000 E; and South (S), coordinates 79.25055556 S, 40.56527778 E (Fig. 1A, Table 1). To minimize contamination, the core sampler was “washed” by drilling multiple 1 m deep holes prior to collecting those for geochemical and microbiological analysis. In addition, the external surface was scrapped and removed before melting the bulk of the cores in a flame-sterilized aluminum pan at controlled temperature (always below 20 °C). Finally, the liquified samples were filtered through 25 mm diameter, 0.22 µm pore size nylon filters in a dedicated small clean area always under a flame. On the other hand, an airborne aerosol sample collector based on a Vaseline retention system was designed (Fig. 1D), built and placed at 1.5 m height at the three N, M, S, locations (Fig. 1A, Supplementary Materials and Methods). The collector was previously sterilized in the lab by wiping exhaustively with 70% alcohol and, after evaporation in laminar flow hood, wrapped with sterile aluminum foil. The sample holders were sterilized similarly and after Vaseline was distributed under laminar flow were carefully included in sterile Whirlpack bags for transportation. A blank control consisting of 25 mm diameter, 0.22 µm filters opened in the mobile laboratory throughout the transect was also included to account for potential human and environmental contamination. Five-day wind back-trajectories were calculated using the Hybrid Single-Particle Lagrangian Integrated Trajectory model (HYSPLIT)49. (Supplementary Materials and Methods for details).
Monitoring deliquescence events
Approximately 300 g of pure NaCl were placed in a squared plastic container. A RH and T sensor and a data logger (U23 Pro V2 from HOBO) were placed inside the salt and set to record data every 15 minutes (Fig. 3A). The box was sealed, and holes were drilled on the top of the plastic container for air circulation and data collected as described in Supplementary Information and in previous works50,51. It was assumed that deliquescent brines could potentially form if two conditions were simultaneously met: TNaCl > -20 °C (the eutectic of saturated NaCl brines) and RHice_NaCl > 75% (the deliquescence point of the pure NaCl). Air relative humidity (RHice_air) and temperature (Tair) were also measured for comparison, although issues with the atmospheric weather station resulted in the loss of data from 27/12/2018 to 05/01/2019.
LDChip and Sandwich immunoassay
LDChip is a 200-antibody microarray designed for detecting a similar number of microbial markers (from peptides to spores) at 1-10 ppb (parts per billion) and 103-104 cells/mL limit of detection for planetary exploration20 (see list of antibodies on LDChip in supplementary Table 5 in52). LDChip was used onboard of the WindSled to analyze the presence of microbial markers in the snow/ice cores by a fluorescence sandwich microarray immunoassay. Briefly, each 1 m-long drill core sample was melted in a sterile pan and filtered through 0.22 µm nylon filter with 25 mm in diameter. Approximately 10 mL of the filtered water samples were allowed to concentrate to 1 mL by heating and evaporation, and then added to a 5 mL tube containing a lyophilized mixture of fluorescence antibodies to obtain a final working dilution of 1/3,000 (2 µg mL-1 of each of the 200 antibodies). Immediately, 50 µL of the mixture were added into one out of nine different incubation chambers of the MultiArray Analysis Module (MAAM) device, each of them with a LDChip53. After incubation and washing, slide chips were read for fluorescence with a portable reader and images were sent by satellite to the science team in Spain for analysis (Supplementary Materials and Methods for details). Each antibody on LDChip is the protein A-purified IgG fraction of its corresponding polyclonal rabbit antiserum from laboratory collection, except two commercial ones (Supplementary Table 3).
DNA extraction, PCR amplification, sequencing, and analysis
Total DNA was extracted from one quarter of filter from each melted snow/ice samples (DN1-4, DM1-4 and DS1-4), and a blank control along the trip (CD_trip) by using the DNeasy PowerBiofilm kit (QIAGEN, Germany) following manufacturer’s instructions. Total DNA from air samples (AN, AM, and AS), and a blank control of the Vaseline system (CA_trip) was extracted with DNeasy kit (QIAGEN). Bacterial community composition of all samples was determined using a bar-coded 16S rRNA amplicon sequencing strategy with primers Bakt 341 F and Bakt 805 R (Supplementary Information)54 targeting the V3-V4 hypervariable region of the 16S rRNA gene and sequenced by Illumina MiSeq™ System (Illumina Inc., San Diego, CA, United States). PCR reactions, and DNA sequence processing and analysis were done following state of the art bioinformatic methods (Supplementary Information Materials and Methods for details). Potential contamination was mitigated by removing those ASVs that were detected in the air and the ice blank controls along the trip (CA_trip and CD_trip), as well as those ASVs identified in the PCR control (CA_PCR). In total, 28% of the sequences were removed for the entire data set of samples to avoid misleading interpretations due to contamination (Supplementary Table 4 and Supplementary Data 2). In addition, singletons (only one ASV) were removed. Therefore, all data screening (elimination of non-bacterial entities, contamination and singletons) resulted in the removal of 74 ASVs accounting for 30% of the quality-filtered sequences. Supplementary information for details.
Culture enrichment for Cyanobacteria
A single piece of filter (stored at -20 °C) was incubated in 50 mL of sterile liquid BG11 medium in a 250 mL flask at room temperature, with 30 µE m-2s-1 of PAR and a photoperiod of 12:12 h. First growth was evident after two and a half weeks. DNA was extracted using the QIAGEN DNeasy UltraClean Microbial Kit according to the manufacturer’s instructions. The 16S rRNA gene was amplified using a combination of primers: 8fw (AGAGTTTGATCATGGCTCAG), 341Fw (CCTACGGGNBGCASCAG), Cya359Fw (GGGGAATYTTCCGCAATGGG), 785Rv (GACTACHVGGGTATCTAATCC), CYA781Rv (GACTACTGGGGTATCTAATCCCATT), and U1541Rv (AAG GAG GTG ATC CAA CC), as previously described34, resulting in a PCR product of 1276 bp in length. See Supplementary Information for amplification details, comparison of the 16S rRNA gene sequence with those in the NCBI database, as well as the construction of the phylogenetic tree.
Stable δ18O isotope determination
Water samples collected from multiple cores along the transect and stored frozen (see Sampling), were used to determine the stable isotope 18O and the ratio δ18O, reported as per mL relative to Standard Mean Ocean Water (SMOW). Water isotopes were analyzed in the Climate Change Institute (CCI) using a Picarro Laser Cavity Ringdown Spectrometer (Model L2130-i) with a high throughput vaporizer using the SMOW standard55. Based on various analyses of internal and international standards, the long-term precision is 0.1‰ (1σ) for δ18O.
Geochemical and Mineralogical Analyses
Anions and low molecular weight organic acids were analyzed by Ion Chromatography (IC) by direct injection of filtered (0.22 µm) melted snow/ice samples into a Metrosep A supp 7-250 column (Metrohm AG, Herisau, Switzerland) with 3.6 mM NaCO3 at a flow rate of 0.7 mL/min as mobile phase (SI Materials and Methods for details).
Statistical analysis
Whenever possible, redundant sampling and determination of different parameters were carried out. Details of data analysis and statistics are described for each technique and measurement in the corresponding section in Supplementary information.
We tested the homogeneity of the 5 day back-trajectories within sampling points with a permutational multivariate analysis of variance tests (PERMANOVA), using the information provided by the DTW56 (Dynamic Time Warping) dissimilarity matrix between all pair of curves. We used DTW with haversine distance between points as data are in Latitude-Longitude coordinates. When the hypothesis of homogeneity is rejected, the multiple comparisons are made with Bonferroni correction with overall significance level α = 0.01. DTW dissimilarity matrix and tests were calculated using the dtw and vegan packages in R57. Bacterial community richness and Shannon-Wiener diversity index (H’) of the air and snow/ice core samples were calculated using the vegan package in R. For beta diversity, Bray Curtis distances between samples were calculated with CANOCO5 v5.04 software (Microcomputer Power, Ithaca, NY, USA) and visualized using a nonmetric dimensional scaling (NMDS) (Supplementary Materials and Methods for details).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw DNA sequence reads generated in this study have been deposited at the NCBI Sequence Read Archive (SRA) under the BioProject ID PRJNA1042783. The 18δO data generated and used in this study are provided in the Supplementary information and also deposited at the Climate Change Institute, University of Maine site https://icecoredata.org/Antarctica.html#WindSled. The new cyanobacterial strain is part of Centro de Astrobiología (CAB), INTA-CSIC, strain collection (available upon request), and is going to be deposited into the DMSZ (Germany) strain collection. The deliquescence dataset generated in this study have been deposited in the Astrobiology Habitable Environment Database, retrieved date Sep 19, 2024 (https://doi.org/10.48667/sy0j-yc84). The ICP-MS data and the fluorescence intensity data from immunoassays are provided in supplementary information. Materials & Correspondence: Victor Parro, [email protected].
References
Hare, F. K. & Rubin, M. J. Studies in Antarctic Meteorology. Geogr. J. 133, 554–555 (1967).
Dalrymple, P. C. A Physical Climatology of the Antarctic Plateau. in Studies in Antarctic Meteorology (ed. Rubin, J.) 195–231 (1966). https://doi.org/10.1029/ar009p0195.
Burton, M. G. Astronomy in Antarctica. A&A Rev. 18, 417–469 (2010).
Chamberlin, R. A., Lane, A. P. & Stark, A. A. The 492 GHz Atmospheric Opacity at the Geographic South Pole. Astrophys J. 476, 428–433 (1997).
Davila, A., Kahre, M. A., Quinn, R. & Des Marais, D. J. The Biological Potential of Present-Day Mars. in Planetary Astrobiology (eds. Meadows, V., Arney, G., Schmidt, B., Des Marais, D. & Dotson, R.) 169–184 (Tucson: University of Arizona Press, 2020).
Braaten, D. A. Direct measurements of episodic snow accumulation on the Antarctic polar plateau. J. Geophys Res Atmospheres 105, 10119–10128 (2000).
Yan, Y. et al. Two-million-year-old snapshots of atmospheric gases from Antarctic ice. Nature 574, 663–666 (2019).
Stoppiello, G. A. et al. Seasonality Is the Main Determinant of Microbial Diversity Associated to Snow/Ice around Concordia Station on the Antarctic Polar Plateau. Biology 12, 1193–1193 (2023).
Michaud, L. et al. Snow Surface Microbiome on the High Antarctic Plateau (DOME C). PLoS ONE 9, e104505 (2014).
Christner, B. C. et al. A microbial ecosystem beneath the West Antarctic ice sheet. Nature 512, 310–313 (2014).
Lopatina, A. et al. Metagenomic Analysis of Bacterial Communities of Antarctic Surface Snow. Front Microbiol 7, 398 (2016).
Antony, R. et al. Microbial communities associated with Antarctic snow pack and their biogeochemical implications. Microbiol Res 192, 192–202 (2016).
Malard, L. A. et al. Spatial Variability of Antarctic Surface Snow Bacterial Communities. Front Microbiol 10, 461 (2019).
Carpenter, E. J., Lin, S. & Capone, D. G. Bacterial Activity in South Pole Snow. Appl Environ. Microbiol 66, 4514–4517 (2000).
Warren, S. G. & Hudson, S. R. Bacterial Activity in South Pole Snow Is Questionable. Appl Environ. Microbiol 69, 6340–6341 (2003).
Darcy, J. L., Lynch, R. C., King, A. J., Robeson, M. S. & Schmidt, S. K. Global Distribution of Polaromonas Phylotypes - Evidence for a Highly Successful Dispersal Capacity. PLoS ONE 6, e23742 (2011).
Archer, S. D. J. et al. Airborne microbial transport limitation to isolated Antarctic soil habitats. Nat. Microbiol 4, 925–932 (2019).
Bodhaine, B. A., Deluisi, J. J., Harris, J. M., Houmere, P. & Bauman, S. Aerosol measurements at the South Pole. Tellus B: Chem. Phys. Meteorol. 38, 223 (1986).
Sheridan, P., Andrews, E., Schmeisser, L., Vasel, B. & Ogren, J. Aerosol Measurements at South Pole: Climatology and Impact of Local Contamination. Aerosol Air Qual. Res 16, 855–872 (2016).
Rivas, L. A. et al. A 200-Antibody Microarray Biochip for Environmental Monitoring: Searching for Universal Microbial Biomarkers through Immunoprofiling. Anal. Chem. 80, 7970–7979 (2008).
Gonzalez, S. et al. Weather Observations of Remote Polar Areas Using an AWS Onboard a Unique Zero-Emissions Polar Vehicle. Bull. Am. Meteorol. Soc. 100, 1891–1895 (2019).
Hughes, K. A. et al. Antarctic environmental protection: Strengthening the links between science and governance. Environ. Sci. Policy 83, 86–95 (2018).
Dibb, J. E., Mayewski, P. A., Buck, C. S. & Drummey, S. M. Beta radiation from snow. Nature 345, 25 (1990).
Favier, V. et al. An updated and quality controlled surface mass balance dataset for Antarctica. Cryosphere 7, 583–597 (2013).
Arthern, R. J., Winebrenner, D. P. & Vaughan, D. G. Antarctic snow accumulation mapped using polarization of 4.3‐cm wavelength microwave emission. J. Geophys Res: Atmospheres 111, D06107 (2006).
Galbán, S., Justel, A., González, S. & Quesada, A. Local meteorological conditions, shape and desiccation influence dispersal capabilities for airborne microorganisms. Sci. Total Environ. 780, 146653 (2021).
KATO, K. Factors controlling oxygen isotopic composition of fallen snow in Antarctica. Nature 272, 46–48 (1978).
Quesada, A. & Vincent, W. F. Cyanobacteria in the Cryosphere: Snow, Ice and Extreme Cold. in Ecology of Cyanobacteria II vol. 9789400738553 387–399 (Springer Netherlands, Dordrecht, 2012).
Jungblut, A. et al. Diversity within cyanobacterial mat communities in variable salinity meltwater ponds of McMurdo Ice Shelf, Antarctica. Environ. Microbiol 7, 519–529 (2005).
Lezcano, M. Á. et al. Comprehensive Metabolic and Taxonomic Reconstruction of an Ancient Microbial Mat From the McMurdo Ice Shelf (Antarctica) by Integrating Genetic, Metaproteomic and Lipid Biomarker Analyses. Front Microbiol 13, 799360 (2022).
Christner, B. C., Kvitko, B. H. & Reeve, J. N. Molecular identification of Bacteria and Eukarya inhabiting an Antarctic cryoconite hole. Extremophiles 7, 177–183 (2003).
Mataloni, G. & Komárek, J. Gloeocapsopsis aurea, a new subaerophytic cyanobacterium from maritime Antarctica. Polar Biol. 27, 623–628 (2004).
Welsh, D. T. Ecological significance of compatible solute accumulation by micro- organisms: From single cells to global climate. FEMS Microbiol Rev. 24, 263–290 (2000).
Azua-Bustos, A. et al. Gloeocapsopsis AAB1, an extremely desiccation-tolerant cyanobacterium isolated from the Atacama Desert. Extremophiles 18, 61–74 (2014).
MAZUR, P. Survival of Fungi after Freezing and Desiccation. in The Fungal Population 325–394 (Elsevier, 1968). https://doi.org/10.1016/B978-1-4832-2744-3.50020−1.
Deming, J. W. & Young, J. N. The role of exopolysaccharides in microbial adaptation to cold habitats. Psychrophiles: From Biodiversity to Biotechnology: Second Edition 259–284 https://doi.org/10.1007/978-3-319-57057-0_12 (2017).
Ewert, M. & Deming, J. Sea Ice Microorganisms: Environmental Constraints and Extracellular Responses. Biol. (Basel) 2, 603–628 (2013).
Dani, K. G. S., Mader, H. M., Wolff, E. W. & Wadham, J. L. Modelling the liquid-water vein system within polar ice sheets as a potential microbial habitat. Earth Planet Sci. Lett. 333–334, 238–249 (2012).
Lugg, D. & Shepanek, M. Space analogue studies in Antarctica. Acta Astronaut 44, 693–699 (1999).
Pyne, S. J. The extraterrestrial Earth: Antarctica as analogue for space exploration. Space Policy 23, 147–149 (2007).
Hecht, M. H. et al. Detection of Perchlorate and the Soluble Chemistry of Martian Soil at the Phoenix Lander Site. Science (1979) 325, 64–67 (2009).
Martín-Torres, F. J. et al. Transient liquid water and water activity at Gale crater on Mars. Nat. Geosci. 8, 357–361 (2015).
Chevrier, V. F., Hanley, J. & Altheide, T. S. Stability of perchlorate hydrates and their liquid solutions at the Phoenix landing site, Mars. Geophys Res Lett. 36, L10202 (2009).
Rivera-Valentín, E. G., Chevrier, V. F., Soto, A. & Martínez, G. Distribution and habitability of (meta)stable brines on present-day Mars. Nat. Astron 4, 756–761 (2020).
Parro, V. et al. SOLID3: A Multiplex Antibody Microarray-Based Optical Sensor Instrument for In Situ Life Detection in Planetary Exploration. Astrobiology 11, 15–28 (2011).
Moreno-Paz, M. et al. Life detection and microbial biomarker profiling with signs of life detector-life detector chip during a mars drilling simulation campaign in the hyperarid core of the atacama desert. Astrobiology 23, 1259–1283 (2023).
McKay, C. P. et al. The icebreaker life mission to mars: a search for biomolecular evidence for life. Astrobiology 13, 334–353 (2013).
Blanc, M. et al. Joint Europa Mission (JEM): a multi-scale study of Europa to characterize its habitability and search for extant life. Planet Space Sci. 193, 104960 (2020).
Stein, A. F. et al. NOAA’s HYSPLIT atmospheric transport and dispersion modeling system. Bull. Am. Meteorol. Soc. 96, 2059–2077 (2015).
Andersen, D. T., McKay, C. P. & Lagun, V. Climate Conditions at Perennially Ice-Covered Lake Untersee, East Antarctica. J. Appl Meteorol. Climatol. 54, 1393–1412 (2015).
McKay, C. P., Balaban, E., Abrahams, S. & Lewis, N. Dry permafrost over ice-cemented ground at Elephant Head, Ellsworth Land, Antarctica. Antarct. Sci. 31, 263–270 (2019).
Sánchez-García, L. et al. Time-integrative multibiomarker detection in Triassic-Jurassic rocks from the atacama desert: relevance to the search for basic life beyond earth. Astrobiology 21, 1421–1437 (2021).
Blanco, Y., Moreno-Paz, M. & Parro, V. Experimental protocol for detecting Cyanobacteria in liquid and solid samples with an antibody microarray chip. J. Vis. Exp. 2017, (2017).
Herlemann, D. P. R. et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579 (2011).
Introne, D. S. UMaine CCI Stable Isotope Laboratory Procedures. UMaine.
Berndt, D. & Clifford, J. Using dynamic time warping to find patterns in time series. AAAIWS’94: Proc. 3rd Int. Conf. Knowl. Discov. Data Min. 398, 359–370 (1994).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna https://www.R-project.org (2022).
Acknowledgements
We thank Maria Paz Martín Redondo for technical assistance with ICP-MS analysis. This research has been funded by grant nos. CTM2016-79741-R (A.J., A.Q., P.M.S., S.G.-H.), RTI2018-094368-B-I00 (V.P.), PID2020-116520RB-I00 (A.J., A.Q., S.G.-H.), PID2021−126746NB-I00 (V.P.), and PGC2021-124362NB-I00 (J.W.) by the Spanish Ministry of Science and Innovation/State Agency of Research MCIN/AEI/FEDER 10.13039/501100011033 and by “ERDF A way of making Europe”. M.A.L was supported by a Juan de la Cierva fellowship (FJC2018- 037246-I) from the MCIN/AEI/ 10.13039/501100011033. The authors acknowledge the computer resources, technical expertise and assistance provided by Centro de Computación Científica at the Universidad Autónoma de Madrid (CCC-UAM). Our thanks to Pablo Sanz from CCC-UAM and Juan Antonio Higuera and Nuria Díaz from SEGAINVEX-UAM. We also thank the Foundation Prince Albert II de Monaco for its support to the campaign.
Author information
Authors and Affiliations
Contributions
V.P. wrote the manuscript; V.P., A.F.-D., A.Q., A.J. designed experiments and analyzed data; M.M.-P., M.G.-V., H.M., performed geochemical and LDChip experiments; M.A.-L., M.R.-N., M.A.-F.-M., P.M.S. performed DNA and cell counting analysis; R.L., Provided the WindSled and led the field Campaign; R.L., I.O., H.M., M.O., performed field work including sample collection and on site experiments; A.A.-B. and J.W. performed the characterization of the cyanobacterium strain; M.P. and P.M. performed water isotopic analysis; S.G.-H. performed meteorological analysis. All authors approved the submitted version. A.J. and A.Q. contributed equally to this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Steven Archer and David Pearce for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Parro, V., Lezcano, M.Á., Moreno-Paz, M. et al. Microbial biogeography along a 2578 km transect on the East Antarctic Plateau. Nat Commun 16, 775 (2025). https://doi.org/10.1038/s41467-025-55997-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-55997-6