Abstract
Genetic mutations in apoptosis-inducing factor (AIF) have a strong association with mitochondrial disorders; however, little is known about the aberrant splicing variants in affected patients and how these variants contribute to mitochondrial dysfunction and brain development defects. We identified pathologic AIF3/AIF3-like splicing variants in postmortem brain tissues of pediatric individuals with mitochondrial disorders. Mutations in AIFM1 exon-2/3 increase splicing risks. AIF3-splicing disrupts mitochondrial complexes, membrane potential, and respiration, causing brain development defects. Mechanistically, AIF is a mammalian NAD(P)H dehydrogenase and possesses glutathione reductase activity controlling respiratory chain functions and glutathione regeneration. Conversely, AIF3, lacking these activities, disassembles mitochondrial complexes, increases ROS generation, and simultaneously hinders antioxidant defense. Expression of NADH dehydrogenase NDI1 restores mitochondrial functions partially and protects neurons in AIF3-splicing mice. Our findings unveil an underrated role of AIF as a mammalian mitochondrial complex-I alternative NAD(P)H dehydrogenase and provide insights into pathologic AIF-variants in mitochondrial disorders and brain development.
Similar content being viewed by others
Introduction
Mitochondrial disorders are rare genetic diseases affecting normal functions of various organs, ultimately leading to developmental, neurological, and metabolic abnormalities. Genetic studies have highlighted a crucial role of an evolutionarily conserved flavoprotein, apoptosis-inducing factor (AIF), in patients with mitochondrial disorders. So far, mutations of at least 26 different residues on AIF have been identified in human patients who develop mitochondria-associated diseases including mitochondrial encephalomyopathy, axonal sensorimotor neuropathy, ataxia, deafness, muscle atrophy, and neurodegeneration1,2,3,4,5,6,7,8,9,10,11,12. In line with human genetics findings, neurodegeneration occurs in the cerebellum and retina during aging in Harlequin mice that harbour a proviral insertion mutation in the intron 1 of Aifm1 leading to about 80% loss of AIF expression13. Moreover, genetic knockout studies from various model organisms, including flies, C. elegans, and mice, support that AIF is essential for maintenance of the stability of mitochondrial respiratory chain complexes and oxidative phosphorylation (OXPHOS) via yet unknown mechanisms14,15,16,17. One caveat of these animal models is that the AIF knockout/knockdown strategy cannot fully depict functions and mechanisms underlying aberrant AIF variants/mutations-triggered mitochondrial disorders in humans, which involve both loss-of-AIF and gain-of-AIF mutant/variant. Thus, an appropriate experimental model with both loss of AIF and gain of aberrant AIF mutants/variants is necessary to advance AIF biology beyond the limitations of prior research.
AIF, encoded by AIFM1 on X chromosome, is ubiquitously expressed across various tissues and organs2,18, and contains N-terminal NADH- and flavin adenine dinucleotide (FAD)-binding motifs and a C-terminal ___domain18,19. AIF protein is synthesized initially as a 67 kDa-precursor in the cytoplasm and then imported into mitochondria, where it is processed to a 62 kDa-mature protein by removal of the mitochondrial localization signal at the N-terminus20. Though a small portion of AIF protein (about 20%) is loosely associated with the outer membrane, AIF protein is primarily located in the intermembrane space of mitochondria with its N-terminal hydrophobic ___domain inserted into the inner membrane21, which creates a technical challenge for expression and purification of full-length mature AIF. As such, most previous AIF structural and functional studies have predominantly utilized truncated “soluble” AIF proteins rather than its mature form, resulting in the inconsistent findings and possibly misconceptions that impede our comprehensive understanding of AIF biology and regulation in mitochondria. For example, AIF (Δ1-120 aa) was initially reported to possess the NAD(P)H oxidase activity, but truncated AIF proteins with Δ1-120 aa, Δ1-100 aa, and Δ1-77 aa, as well as His-tagged AIF (Δ1-53 aa) were later found to exhibit no or little NADPH or NADH oxidase activity compared to yeast NADH dehydrogenase NDI1, unless it was reconstituted with complicated bacterial or mitochondrial membrane fractions22,23,24. Besides these conflicting observations, the specific role of AIF in mitochondrial respiration and redox homeostasis also appears to be controversial25,26,27,28. AIF-deficient Harlequin mice display increased oxidative stress and neurodegeneration in the cerebellum during aging, indicating that AIF may function as a free-radical scavenger13,29. In contrast, other studies showed that AIF might be a ROS producer22,26,30. NDI1 was recognized as a type II dehydrogenase and an alternative substitute for mitochondrial complex I in yeast31,32, however, it remains unknown whether AIF acts as an alternative NAD(P)H dehydrogenase for the mammalian mitochondrial complex I.
Previous studies identified several AIF splicing isoforms, including a brain-specific AIF2 isoform with a more hydrophobic mitochondrial inner membrane sorting signal encoded by an alternative exon 2b rather than exon 2a33, and three short AIF isoforms (AIFsh, AIFsh2 and AIFsh3) with unknown biological functions18,34,35. We recently characterized another splicing isoform named as AIF3, which is induced under pathological conditions, including ischemic injury in brain tissues from both humans and mice36. AIF3 lacks the AIFM1 exons 2 and 3-encoded hydrophobic membrane-insertion ___domain and matrix processing peptidase cleavage site M53↓A54 critical for AIF maturation. AIF3 splicing causes notable mitochondrial disorganization and neurodegeneration in targeted mouse brain regions36.
In this study, we screened AIF variants in postmortem brain tissues of pediatric individuals with mitochondrial disorders and analyzed whole exome sequencing data from 141, 456 human samples. We revealed that RNA splicing vulnerability at exon 2 and genetic/somatic mutations enriched at exon 3 of AIFM1 robustly increased risks of pathologic AIF3 and AIF3-like (AIF3L) splicing, which provides a fundamental mechanistic support for our recently established AIF3 splicing mouse model36. Using in vitro tag-free mature AIF-based biochemical assays, cellular loss-of-function and gain-of-function studies, and AIF3 splicing models in vivo, we further found that full-length mature AIF was a mammalian mitochondrial complex I alternative NAD(P)H dehydrogenase and possessed previously unrecognized glutathione reductase (GR) activity, whereas AIF3 lost GR and NAD(P)H dehydrogenase activities. AIF3 splicing disrupted the mitochondrial complex assembly and glutathione (GSH)-redox homeostasis causing mitochondrial dysfunction and brain development deficits, which could be rescued partially by expression of NDI1. Together, this study revealed the fundamental role of AIF in mitochondria and shed light on the detrimental effects and mechanisms of AIF3 splicing in mitochondrial disorders.
Results
Genetic variants and mutations are the potential risk factors contributing to AIF3 splicing in human brain with mitochondrial disorders
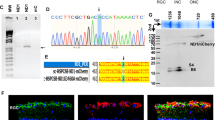
To determine whether AIF3 splicing is induced in the postmortem frontal cortex tissues from pediatric individuals with mitochondrial disorders (Table S1), we performed PCR with primers specifically binding to the AIF3 splicing junction sequence (hAIF-Fw2E1/4, Key Resources Table), and detected AIF3 mRNA in all seven patients but not their age-matched controls (Fig. 1a, b). The cellular expression of AIF3 splicing was further examined in the cortex and hippocampus of these postmortem human brain tissues by RNA in situ hybridization with the probe specifically recognizing AIF3 (hAIF3-3-SP6-Fw/hAIF3-3-T7-Re, Key Resources Table). The AIF3+ cell number was increased in both cortex and hippocampus of all 7 pediatric individuals with mitochondrial disorders (Fig. 1c, d, and Supplementary Fig. 1a). The specificity of AIF and AIF3 RNA probes was verified in human cell line MDA-MB-231 expressing endogenous AIF or CRISPR/Cas9-produced AIF3 (Supplementary Fig. 1b). Likewise, mouse AIF3 RNA in situ was demonstrated in brain tissues from AIF3 splicing mice and AIF3 transgenic (AIF3 Tg) mice, but not their littermate controls (Supplementary Fig. 1c). AIF3 protein levels were elevated in the postmortem frontal cortex from pediatric individuals with mitochondrial disorders, whereas it was barely detectable in normal brain tissues from age-matched controls (Fig. 1e, f). These findings indicate that AIF3 splicing is enhanced in pediatric individuals with mitochondrial disorders.
a, b Expression (a) and quantification (b) of AIF3 mRNA in the postmortem frontal cortex tissues from pediatric individuals with mitochondrial disorders using AIF3-specific primers. Data are shown as mean ± S.E.M throughout this figure unless otherwise indicated, n = 3 for control (CTL) and 7 for patients. **p = 0.0099, by unpaired 2-tailed Student’s t test. c, d In situ hybridization for AIF3 mRNA (c) and quantification (d) in the cortex and hippocampus of pediatric individuals with mitochondrial disorders and normal controls using AIF3-specific probe. Scale bar, 10 μm. n = 3 for controls and 7 for patients. *p = 0.045; **p = 0.0022; ****p < 0.0001, by one-way ANOVA with Dunnett’s multiple comparison test. e, f Expression (e) and quantification (f) of AIF3 protein in the frontal cortex of pediatric individuals with mitochondrial disorders. n = 3 for controls and 7 for patients. *p = 0.0132 by unpaired 2-tailed Student’s t test. g, h Expression (g) and Sanger sequencing (h) of AIF mRNA variants in the postmortem frontal cortical tissues from individuals without neurological diseases. i Percentage of ΔE2-AIF/total AIF in human frontal cortex. Group 1, ΔE2-AIF- (n = 7); Group 2, ΔE2-AIF+ (n = 8). ****p < 0.0001, by unpaired 2-tailed Student’s t test. j Frequency of WT and ΔE2-AIF in the human frontal cortex. n = 15 for normal and 7 for patients. k Mutation frequency of each exon of AIFM1 was analyzed based on the total allele counts of missense, synonymous, and splicing variants in each exon across 141,456 populations using whole exome sequencing. Data were retrieved from Exome Variant Server (https://gnomad.broadinstitute.org/gene/ENSG00000156709?dataset=gnomad_r2_1). l–n Expression of AIF variants at mRNA (l), protein (m) and its protein quantification (n) induced by CRISPR/Cas9-based genome editing in MDA-MB-231 cells. n = 3 independent biological replicates. ****p < 0.0001, by one-way ANOVA with Tukey’s multiple comparison test. o 5’-RACE analysis of AIF splicing variants in the postmortem frontal cortical tissues from 7 pediatric individuals with mitochondrial disorders.
To identify possible genetic variations at the 5’ end of AIFM1 where AIF3 is spliced, we screened AIF mRNA products in 15 human postmortem brain tissues who had no obvious neurological deficits (Table S2) using human AIF primers (hAIF-Fw128 and hAIF-Re761, Key Resources Table). In addition to a dominant wildtype (WT) AIF band, a smaller AIF PCR product lacking exon 2 (ΔE2) was detected and confirmed by Sanger sequencing in 8 out of 15 human brain tissues, although its expression levels were quite low ranging from 2% to 7% relative to the total AIFM1 mRNA (Fig. 1g–i). The frequency of ΔE2AIF mRNA was about 50% in normal human brain tissues, but was increased to 100% in pediatric individuals with mitochondrial disorders (Fig. 1j). Next, we analyzed AIFM1 genetic variations identified by whole exome sequencing from 141, 456 human samples in the Exome Variant Database (https://gnomad.broadinstitute.org/gene/ENSG00000156709?dataset=gnomad_r2_1) and found a much higher genetic mutation rate (63%) in exon 3 (Fig. 1k, see AIFM1 exon mutation list in Source Data). These findings indicate that AIF lacking exon 2 is an mRNA splicing variant in about 50% of people with a low occurrence, and that exon 3 of AIFM1 is highly susceptible to genetic mutations in humans.
Next, we asked whether genetic or acquired somatic mutations in AIFM1 exon 3 in human populations carrying ΔE2AIF variant increase the incidence of AIF3 splicing. To this end, CRISPR/Cas9-based single-guide RNAs (sgRNAs) were designed to target exon 2 or 3 of human AIFM1, aiming to generate identical or similar mutations at the specific sites, including Ser57, Leu69, Thr77, Gly101, and Arg98, as observed in human populations according to the Broad Institute Database (Supplementary Fig. 1d, Source Data). We found that sgRNA3 targeting exon 2 of AIFM1 (E2g3) induced mRNA variant lacking exon 2 (ΔE2AIF) and sgRNA4 targeting exon 3 of AIFM1 (E3g4) resulted in a low incidence of AIF3 splicing in MDA-MB-231 cells (Fig. 1l). sgRNA3 and sgRNA4 combo robustly increased the incidence of AIF3 splicing (Fig. 1l). To confirm AIF3 protein induction, we customized AIF3-1157/1159 antibodies using the junction region encoded by exons 1 and 4 of human AIFM1 as an immunogen, which specifically recognized AIF3, but not WT AIF, both of which were immunoprecipitated by a commercially available anti-AIF E1 antibody (Supplementary Fig. 1e). AIF-147 antibody produced from the immunogen encoded by exon 3 of human AIFM1 detected WT AIF but not AIF3 (Supplementary Fig. 1e). A notable increase in AIF3 protein expression was observed in MDA-MB-231 cells transduced with both sgRNA4 and sgRNA3, while the expression of WT AIF protein was significantly reduced in these transduced cells (Fig. 1m, n). Similarly, sgRNAs targeting exon 3 of mouse Aifm1 increased AIF3 splicing at both mRNA and protein levels, which could be detected by AIF3-specific PCR and antibody (Supplementary Fig. 1f–j). However, sgRNAs targeting exon 2 of mouse Aifm1 induced AIF3 splicing at a very low level, because AIF3 was detected by AIF3-specific PCR and anti-AIF3-1157 antibody only after protein immunoprecipitation (Supplementary Fig. 1f–h, k). These results indicate that combined genetic or somatic mutations in exons 2 and 3 of AIFM1 increase the risk of AIF3 splicing.
To further confirm AIF variants in pediatric brain with mitochondrial disorders (Table S1), 5’ RACE coupled with TA-cloning and Sanger sequencing was performed. AIF3 and ΔE2AIF splicing isoforms besides WT AIF were identified in all 7 patients (Fig. 1o and Supplementary Fig. 1l). Moreover, the high frequency of multiple AIF3L isoforms, producing similar mature AIF3 proteins but initiating at different sites including i) Cys24 at the end of exon 1 but lacking exons 2-3, ii) Met88, Tyr95, Lys109 or Lys111 at the end of exon 3, or iii) Pro124 or His131 at the beginning of the exon 4, was identified in postmortem tissues from pediatric individuals with mitochondrial disorders (Fig. 1o and Supplementary Fig. 1l). In addition, a few other short isoforms or mutant isoforms were identified as indicated in Fig. 1o. These findings suggest that genetic/somatic mutations and variants under pathological conditions like mitochondrial disorders increase the risk of AIF3(L) splicing in humans.
AIF3 splicing causes mitochondrial abnormalities and mouse brain development deficits
To investigate whether AIF3 is localized to mitochondria, the subcellular localization of AIF3 was determined by the subcellular fragmentation study. Both AIF and AIF3 proteins were detected only in the mitochondrial fraction, but not the nuclear or cytosol fraction (Supplementary Fig. 2a). Histone H3, CytC, and α-Tubulin were used as the specific markers for the nuclear, mitochondrial, and cytosol fractions, respectively. To further examine the sub-mitochondrial localization of AIF3 in the cell, mitochondrial outer membrane, intermembrane space, inner membrane, and matrix fractions were isolated from the cortex of WT and AIF3 Tg mice. We found that both AIF and AIF3 were mainly associated with the inner membrane, while a small portion of proteins were associated with the outer membrane (Fig. 2a). AIF3 but not AIF was also detected in the matrix (Fig. 2a). Similar results were observed in MEFs (Supplementary Fig. 2b). The localization of AIF3 in mitochondria was further confirmed by immunostaining with anti-AIF-E1 antibody and MitoTracker. Endogenous AIF or AIF3 was colocalized with MitoTracker in MEFs and cortical neurons (Fig. 2b). Collectively, these findings indicate that the endogenously spliced AIF3 protein is primarily located in inner and outer mitochondrial membranes with partial mislocalization to the mitochondrial matrix.
a Submitochondrial localization of AIF and AIF3 in mouse brain. Mitochondria isolated from WT and AIF3 Tg mouse brain were fractionated into outer membrane (OM), intermembrane space (IMS), inner membrane (IM) and matrix (Mx). n = 5 independent biological replicates. b Mitochondrial morphology and subcellular localization in WT, AIF3 splicing, AIF3-AIF (rescue), OE-AIF, OE-AIF3, and AIF KO MEFs and cortical neurons by immunostaining with anti-AIF-E1 antibody (red) and MitoTracker (green). Representative images were from three independent experiments. Scale bar, 10 μm. c Quantification of mitochondrial area distribution in the cytosol (as 100%) shown in (b). Data are shown as mean ± S.E.M throughout this figure. n = 5 independent biological replicates. *p = 0.0296, ****p < 0.0001, by one-way ANOVA with Tukey’s multiple comparison test. d, e Quantification of donut-shaped mitochondria in MEFs (d) and cortical neurons (e) shown in b. n = 10 biological replicates. *p = 0.0468, ****p < 0.0001, by one-way ANOVA with Tukey’s multiple comparison test (d) and unpaired 2-tailed Student’s t test (e). f Mitochondrial ultrastructure in WT and AIF3 splicing MEFs and cortical neurons, and cortex of WT, AIF3 splicing, and AIF3 transgenic (Tg) mice by electron microscopy. g–j Percentage of abnormal mitochondria with disorganized/thicker cristae in MEFs (g), neurons (h), cortex of AIF3 splicing (i) and AIF3 Tg (j) mice shown in f. n = 5 independent biological replicates. ****p < 0.0001, by unpaired 2-tailed Student’s t test. k Nissl staining of hippocampus of AIF3 splicing mice (Emx1-Cre + ) and their littermate controls at E15, P0, and P120. n = 6 mice. Scale bar, 200 μm. l Immunostaining of NeuN+ neurons in AIF3 splicing mice (Emx1-cre + ) and their littermate controls. n = 6 mice. Scale bar, 200 μm.
Notably, mitochondria were accumulated surrounding the nucleus with a donut-shaped morphology in AIF3 splicing MEFs and cortical neurons, which was distinct to spindle-like shaped mitochondria evenly distributed in the entire cytosol in WT MEFs (Fig. 2b–e). Re-expression of AIF did not prevent altered mitochondrial distribution and morphological changes conferred by AIF3 splicing (Fig. 2b–e). Overexpression of AIF or AIF3 alone had no obvious effect on mitochondrial morphology in MEFs (Fig. 2b–e). Likewise, knockout (KO) of AIF did not alter mitochondrial distribution, although it increased the length of spindle-like mitochondrial structure. We further studied the effect of AIF3 splicing on mitochondrial morphological changes by electron microscopy. Mitochondria were disorganized in AIF3 splicing cells as reflected by the vacuoles or disrupted cristae structures, which were observed not only in MEFs and cortical neurons in vitro but also in the cortex of AIF3 splicing mice (Fig. 2f–j). Although gain-of-AIF3 in the presence of AIF did not alter overall mitochondrial structure, the mitochondrial cristae displayed a condensed appearance in the cortex of AIF3 Tg mice (Fig. 2f, j), similar to those observed from AIF conditional KO in liver as reported previously16. These findings indicate that AIF3 is partially mislocalized to the mitochondrial matrix and alters mitochondrial morphology, structure and distribution.
To study the effect of AIF3 splicing on mouse embryonic and postnatal brain development, AIF3 splicing was induced during the embryonic stage in the forebrain and whole brain by crossing AIFfl/fl mice with transgenic Emx1-Cre and Nestin-Cre mouse lines, respectively. We found that Emx1-Cre + -inducible AIF3 splicing mice had reduced body size, notably the brain, and hair loss on the cheek altering facial appearance (Supplementary Fig. 2c). The thickness of brain cortex was reduced in AIF3 splicing mice, which was further supported by Nissl staining and immunofluorescence staining of CUX1 (layer II and III marker) and TBR1 (layer VI marker) (Supplementary Fig. 2d–k). Furthermore, AIF3 splicing impaired hippocampal development at embryonic day 15 (E15), postnatal day 0 (P0), and P120 (Fig. 2k, l, and Supplementary Fig. l–o). Nestin-Cre was expressed in the whole brain to induce AIF3 splicing, which reduced the brain size and cell density in the cortex and hippocampus leading to early death before P10 (Supplementary Fig. 2p, q). AIF3 splicing induced postnatally by CamkIIα-iCre has no effect on brain morphogenesis but causes early neurodegeneration leading to mouse death by P12036. These findings indicate that AIF3 splicing disrupts mitochondria leading to deficits in brain development during the embryonic stage.
AIF3 splicing impairs mitochondrial functions and activities of respiratory chain complexes
To study the effect of AIF3 splicing on mitochondrial functions, we first assessed the mitochondrial membrane potential in both WT and AIF3 splicing MEFs using tetramethylrhodamine, ethyl ester (TMRE) fluorescent dye. AIF3 splicing significantly decreased the mitochondrial membrane potential in MEFs, which could not be restored by re-expression of AIF (Fig. 3a and Supplementary Fig. 3a). Interestingly, overexpression of AIF or AIF3 alone exhibited no obvious effect on the mitochondrial membrane potential in WT MEFs (Fig. 3a and Supplementary Fig. 3a). The uncoupler of mitochondrial OXPHOS, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP, 1 μM), was applied to induce loss of mitochondrial membrane potential as a positive control. These findings support that AIF3 splicing impairs mitochondrial membrane potential.
a Analysis of mitochondrial membrane potential by TMRE staining in WT, AIF3 splicing, OE-AIF, OE-AIF3, and AIF3-AIF (rescue) MEFs. Representative images were from three independent experiments. Scale bar, 100 μm. b Analysis of total cellular ATP levels in WT, AIF3 splicing, OE-AIF, OE-AIF3, and AIF3-AIF (rescue) MEFs. Data are shown as mean ± S.E.M throughout this figure, n = 12 biological replicates. **p = 0.0014, by one-way ANOVA with Tukey’s multiple comparison test. c Analysis of mitochondrial ATP levels with 2-DG (5 mM) in WT, AIF3 splicing, OE-AIF, OE-AIF3, and AIF3-AIF (rescue) MEFs. n = 15 (WT/AIF3/AIF3-AIF) and 20 (OE-AIF/OE-AIF3) biological replicates. ****p < 0.0001, by one-way ANOVA with Tukey’s multiple comparison test. d Analysis of OCR in WT, AIF3 splicing, AIF3-AIF (rescue), OE-AIF, and OE-AIF3 MEFs. n = 4 independent biological replicates. O, Oligomycin. F, FCCP. A, antimycin A. R, rotenone. e–i Enzymatic activities of mitochondrial complexes I to V in WT, AIF3, AIF3-AIF (rescue), OE-AIF, and OE-AIF3 MEFs. n = 20 (WT/AIF3), 16 (AIF3-AIF), 5 (OE-AIF) and 4 (OE-AIF3) (e); n = 6 (f); n = 9 (WT/AIF3/AIF3-AIF) and 6 (OE-AIF/OE-AIF3) (g); n = 9 (WT/AIF3/AIF3-AIF) and 8 (OE-AIF/OE-AIF3) (h); n = 8 (WT/AIF3/AIF3-AIF) and 6 (OE-AIF/OE-AIF3) (i) biological replicates. ****p < 0.0001, by one-way ANOVA with Tukey’s multiple comparison test. j–n Enzymatic activities of mitochondrial complexes I to V in WT and AIF3 splicing (Emx1-Cre + ) mouse brain. n = 6 mice (WT/AIF3) (j), 7 mice (WT) and 6 mice (AIF3) (k–n). **p = 0.0039, ***p = 0.0005, ****p < 0.0001, by unpaired two-tailed Student’s t test. o–r In gel activity assay in mitochondria isolated from MEFs and forebrain of AIF3 splicing (Emx Cre + ) and AIF3 Tg mice. SC, supercomplex. C, complex.
Next, the luminescence assay was conducted to assess ATP production in both WT and AIF3 splicing MEFs in the presence of glucose (10 mM) with or without a glycolysis inhibitor 2-deoxy-d-glucose (2-DG, 5 mM). The total cellular ATP levels in AIF3 splicing MEFs were comparable to those in WT MEFs (Fig. 3b). Interestingly, overexpression of AIF3 increased the total cellular ATP levels in WT MEFs (Fig. 3b). However, when glycolysis was inhibited by 2-DG, ATP production significantly decreased in AIF3 splicing MEFs compared to WT MEFs (Fig. 3c). Re-introduction of AIF failed to restore mitochondria-derived ATP production in AIF3 splicing MEFs (Fig. 3c). Moreover, overexpression of AIF3 resulted in a partial reduction in mitochondria-derived ATP production in WT MEFs (Fig. 3c). These findings support that AIF3 splicing reduces mitochondrial ATP production but enhances glycolysis-derived ATP production.
The effect of AIF3 splicing on OXPHOS was further evaluated by measurement of the oxygen consumption rate (OCR) in MEFs in the presence of mitochondrial ATP synthesis inhibitor oligomycin (1 μM), the potent uncoupler of mitochondrial OXPHOS FCCP (1 μM), mitochondrial complex I inhibitor rotenone (1 μM), and mitochondrial electron transport inhibitor antimycin A (5 μM) using a Seahorse Bioscience XF-24 Extracellular Flux Analyzer. The basal OCR was significantly decreased in AIF3 splicing MEFs compared to WT MEFs (Fig. 3d and Supplementary Fig. 3b). The ATP-linked OCR, proton leak OCR, maximal OCR, reserve capacity, and non-mitochondrial OCR all were decreased in AIF3 splicing MEFs (Fig. 3d and Supplementary Fig. 3c–g). Re-expression of AIF did not rescue the OCR in AIF3 splicing MEFs, although it slightly increased the basal OCR (Fig. 3d and Supplementary Fig. 3b–g). Overexpression of AIF3 decreased the ATP-linked respiration, maximal respiratory capacity, and non-mitochondrial OCR in WT MEFs, whereas overexpression of AIF failed to do so (Fig. 3d and Supplementary Fig. 3b–g). These findings support an inhibitory role of AIF3 splicing in mitochondrial respiration and ATP production.
To understand the molecular mechanism underlying AIF3 splicing-induced mitochondrial dysfunction, we studied the activities of respiratory complexes in mitochondria isolated from AIF3 splicing and WT MEFs. As shown in Fig. 3e–i, the activities of complexes I, III, IV and V were significantly decreased in AIF3 splicing MEFs compared to WT MEFs, whereas the activity of complexes II remained unchanged. Expression of AIF partially rescued the activities of complexes III and V, but not complexes I and IV, in AIF3 splicing MEFs (Fig. 3e–i). Overexpression of AIF3 alone decreased activities of complexes IV and V in WT MEFs, whereas AIF overexpression had no effect (Fig. 3e–i). In line with in vitro observations, the activities of mitochondrial respiratory complexes I, III, IV and V were significantly reduced in the cortex and hippocampus of both Emx1-Cre+ and CamkIIα-iCre-inducible AIF3 splicing mice (Fig. 3j–n and Supplementary Fig. 3h–l). Only the mitochondrial complex V activity was modestly decreased in AIF3 Tg mice (Supplementary Fig. 3m–q).
We further assessed mitochondrial complexes by blue native polyacrylamide gel electrophoresis (BN-PAGE), showing that mitochondrial complex I was reduced in AIF3 splicing MEFs and mouse brain (Supplementary Fig. 3r). In-gel activity assay for mitochondrial complexes I, II, IV and V was also performed in AIF loss-of-function, AIF or AIF3 gain-of-function, and AIF3 splicing MEFs and mice. Mitochondrial complexes I, IV and V were reduced in AIF3 splicing MEFs and mice, which could not be rescued by re-expressing AIF in AIF3 splicing cells (Fig. 3o–r, Supplementary Fig. 3s–v and quantification in Source Data), consistent with our results above (Fig. 3e–n and Supplementary Fig. 3h–l). Loss-of-AIF alone reduced the in-gel activity of complexes I and V, but not complexes II and IV (Fig. 3o–r). We also confirmed that overexpression of AIF3 reduced the complex IV and V activities in WT MEFs (Fig. 3q, r) and that the complex II activity was not affected by overexpression of AIF3 or AIF (Fig. 3p). Taken together, these data indicate that AIF3 splicing reduces mitochondrial complex activities leading to mitochondrial dysfunction.
AIF3 splicing causes mitochondrial complex disassembly and increases ROS levels
To further elucidate the mechanism underlying AIF3 splicing-reduced mitochondrial complex activity, we examined the effect of AIF3 splicing on the stability of mitochondrial complexes. AIF3 splicing reduced the protein levels of NDUFA9, NDUFS3, NDUFA13, ND1, and NDUFB8 of the complex I, CytB of the complex III, MTCO1 of the complex IV in MEFs, whereas loss of AIF diminished NDUFB8 and MTCO1 only (Fig. 4a, quantification in Source Data). Re-expression of AIF failed to restore NDUFS3, ND1, CytB, and MTCO1 protein levels in AIF3 splicing MEFs, although NDUFA9, NDUFA13, and NDUFB8 protein levels were partially rescued (Fig. 4a, quantification in Source Data). Overexpression of AIF3 alone did not affect the expression of mitochondrial complex subunits except for MTCO1 in WT MEFs (Fig. 4a, quantification in Source Data). Reduced expression of NDUFS3 and NDUFA13 was further confirmed by immunostaining in AIF3 splicing MEFs (Supplementary Fig. 4a, quantification in Source Data). Similar results were observed in AIF3 splicing cortical neuron cultures and mouse brains (Fig. 4b and Supplementary Fig. 4b, c, quantification in Source Data). Again, AIF3 overexpression (AIF3 Tg) did not alter the expression of mitochondrial complex subunits in mouse brain (Supplementary Fig. 4c, quantification in Source Data). Interestingly, loss- or gain-of-AIF or AIF3 splicing had no effect on many other non-mitochondrial complex proteins (Supplementary Fig. 4d, quantification in Source Data). These findings indicate that AIF3 splicing specifically causes mitochondrial complex disassembly.
a Immunoblot analysis of mitochondrial complex subunits in WT, AIF KO, AIF3 splicing, AIF3-AIF (rescue), OE-AIF, and OE-AIF3 MEFs. n = 3 independent biological replicates. b Immunoblot analysis of complex I subunits in AIF3 splicing mice (Nestin-Cre + ) and their littermate control. n = 3 independent biological replicates. c Mitochondrial DNA mass in WT, AIF3 splicing, and AIF3-AIF (rescue) MEFs. Data are shown as mean ± S.E.M throughout this figure, n = 3 independent biological replicates. *p = 0.0269 (WT vs. AIF3), *p = 0.0103 (WT vs. AIF3-AIF), by one-way ANOVA with Tukey’s multiple comparison test. d Volcano plot showing differentially expressed genes in AIF3 splicing MEFs compared with WT MEFs. n = 2 independent RNA-seq replicates. The p value was calculated by the edgeR package. e Hallmark pathways with Gene Set Enrichment Analysis of differentially expressed genes in AIF3 splicing MEFs. NES, normalized enrichment score. f, g Enrichment plot of OXPHOS and ROS pathways in WT vs. AIF3 splicing MEFs. h, i Representative images (h) and quantification (i) of mitochondrial ROS levels by MitoSOX staining in WT and AIF3 splicing MEFs treated with DMSO or H2O2 (250 μM). Scale bar, 100 μm. n = 30 biological replicates. ****p < 0.0001, by two-way ANOVA with Tukey’s multiple comparison test. j Analysis of mitochondrial ROS levels by MitoSOX before, during, and after H2O2 (200 μM) stimulation in WT and AIF3 splicing MEFs with or without GR overexpression. n = 3 independent biological replicates. *p = 0.0388; **p = 0.0038, 0.0049, 0.0012 in time order; ****p < 0.0001, by two-way ANOVA with Tukey’s multiple comparison test. k Analysis of mitochondrial ROS levels by MitoSOX before, during, and after H2O2 stimulation (200 μM) in GRKO WT and AIF3 splicing MEFs. n = 3 independent biological replicates. ****p < 0.0001, by two-way ANOVA with Tukey’s multiple comparison test.
To determine whether AIF3 splicing induces mitochondrial complex disassembly at the DNA or RNA level, mitochondrial DNA mass and bulk RNA-seq were performed. We found that AIF3 splicing significantly increased mitochondrial DNA mass in MEFs, which was not reversed by re-expression of AIF (Fig. 4c). Likewise, AIF3 splicing increased mRNA expression of ND1, ND2, ND3, and COX2 but not other mitochondrial-encoded subunits in MEFs, which was not reversed by re-expression of AIF (Supplementary Fig. 4e). Overexpression of AIF or AIF3 alone had no effect on mRNA expression of 13 mitochondrial-encoded subunits (Supplementary Fig. 4e). Bulk RNA-seq detected 2327 differentially expressed genes in AIF3 splicing MEFs (Fig. 4d), which were enriched in pathways including ROS, OXPHOS, glycolysis, and DNA repair (Fig. 4e–g). Re-expression of AIF did not alter upregulation of genes involved in OXPHOS in AIF3 splicing MEFs (Supplementary Fig. 4f). However, AIF3 splicing had no effect on the mRNA expression of nuclear-encoded mitochondrial complex subunits in MEFs (Supplementary Fig. 4g, h). These findings exclude a role of transcriptional regulation in AIF3 splicing-induced mitochondrial complex subunit instability.
Considering that AIF3 splicing caused mitochondrial complex disassembly and induced genes involved in OXPHOS and ROS pathways, we next examined the effect of AIF3 splicing on cellular ROS levels in WT and AIF3 splicing MEFs in the presence or absence of H2O2 treatment (250 μM, 2 h) using MitoSOX staining. The overall basal mitochondrial ROS levels were not changed between WT and AIF3 splicing MEFs (Fig. 4h, i). However, upon 2 h H2O2 stimulation followed by a 6 h-recovery period, mitochondrial ROS levels were significantly increased in AIF3 splicing MEFs compared to WT MEFs, which could not be reversed by re-expression of AIF (Fig. 4h, i).
To assess whether AIF3 splicing-induced ROS accumulation was attributed to the increase in ROS generation or the decrease in ROS clearance, we monitored ROS levels in WT and AIF3 splicing MEFs before, during H2O2 treatment (250 μM, treatment duration for 5 min, 10 min, 1 h or up to 2 h), and after an additional 6-h recovery. WT and AIF3 splicing MEFs overexpressing GR, which controls GSH regeneration for ROS clearance, served as controls. Before H2O2 treatment, the basal levels of ROS had little difference between WT and AIF3 splicing MEFs with or without GR expression (Fig. 4j). Upon H2O2 treatment, ROS levels rapidly increased in WT MEFs, reaching a peak within 5-10 min (Fig. 4j). Subsequently, they partially declined within 30-120 min even in the presence of H2O2 before returning to the basal levels after a 6 h-recovery period (Fig. 4j). AIF3 splicing significantly increased ROS levels compared to WT MEFs after 5 and 10 min-H2O2 treatment. Although they were also partially reduced during 6-h ROS clearance phases in AIF3 splicing MEFs, ROS levels were significantly higher than those in WT MEFs and failed to return to the basal levels (Fig. 4j). H2O2-induced ROS was not evident in MEFs overexpressing GR, indicating that ROS clearance occurs very rapidly within minutes. In parallel, to eliminate the interference from the ROS clearance process, GR KO in WT and AIF3 splicing MEFs were conducted, allowing us to determine whether AIF3 splicing increases ROS generation. As expected, GR KO increased the basal levels of ROS by ~1.5-fold in MEFs, and H2O2 treatment had a modest impact on increase in ROS levels in these cells (Fig. 4k). However, a robust increase in ROS levels was observed in GR-deficient AIF3 splicing MEFs (AIF3-GRKO) at 1 h after H2O2 treatment, which was well maintained during the 6-h recovery period (Fig. 4k). GR overexpression and KO in WT and AIF3 splicing MEFs were confirmed by immunoblot assay (Supplementary Fig. 4i). These findings indicate that AIF3 splicing not only increases ROS generation but also suppresses ROS clearance.
AIF3 splicing impairs AIF’s NADPH and NADH dehydrogenase activities
Given the importance of NAD(P)H dehydrogenase in ROS production within the mitochondrial respiratory chain and in cellular antioxidant defense, we comprehensively characterized these biochemical properties of AIF/AIF3 to explore the molecular mechanisms underlying AIF3 splicing-disrupted ROS homeostasis. Our recent study showed that AIF3 exhibits a significantly weaker NADH dehydrogenase activity than AIF at the same concentrations36. Notably, AIF but not AIF3 possessed a previously unrecognized NADPH dehydrogenase activity as shown by in vitro biochemical assay (Fig. 5a). The cellular metabolomic analysis revealed increased NADPH levels and decreased NADP+/NADPH ratio in AIF3 splicing MEFs, which mimicked the phenotypes of AIF KO but failed to be rescued by re-expression of AIF (Fig. 5b–d and Supplementary Fig. 5a–c). Similarly, the ratio of NAD + /NADH was reduced in AIF3 splicing MEFs (Supplementary Fig. 5d–f). The homeostasis of NAD+/NADH and NADP+/NADPH was further studied in the cortex and hippocampus from AIF3 splicing mice induced by Emx1-Cre. In line with in vitro results above, the cellular levels of NADH and NADPH were elevated in AIF3 splicing mouse brain, whereas NAD+ and NADP+ levels and ratios of NAD+/NADH and NADP+/NADPH were reduced (Fig. 5e–j). These findings indicate that AIF3 lacks the NADH/NADPH dehydrogenase activity and that AIF3 splicing disrupts NAD+/NADH and NADP+/NADPH homeostasis.
a Analysis of the NADPH dehydrogenase activity of purified AIF and AIF3 (3 μM). Data are shown as mean, n = 4 independent biological replicates. b–d Analysis of NADPH, NADP+, and NADP+/NADPH ratio in WT, AIF3 and AIF3-AIF (rescue) MEFs. Data are shown as mean ± S.E.M throughout this figure unless otherwise indicated, n = 5 independent biological replicates. **p = 0.0014 and ****p < 0.0001 (b); *p = 0.0155 and **p = 0.0022 (d); by one-way ANOVA with Tukey’s multiple comparison test. e–g Analysis of NADH, NAD+, and NAD+/NADH ratio in the hippocampus of WT and AIF3 splicing (Emx1-Cre + ) mice. n = 10 biological replicates. **p = 0.0012; ****p < 0.0001, by unpaired 2-tailed Student’s t test. h, j Analysis of NADPH, NADP+, and NADP+/NADPH ratio in the hippocampus of WT (n = 9) and AIF3 splicing (Emx1-Cre + , n = 7) mice. ****p < 0.0001, by unpaired 2-tailed Student’s t test. k Analysis of the NADPH dehydrogenase activity of purified AIF and its mutants. Data are shown as mean, n = 4 independent biological replicates. l Analysis of the NADPH dehydrogenase activity of purified AIF and its deletion mutants (3 μM). Data are shown as mean, n = 4 (Auto/AIF/AIF-Δ3), n = 6 (AIF-Δ2) biological replicates. m Immunoblot analysis of purified AIF and AIF3 monomer and/or dimer (7.5-30 ng) using anti-AIF-E1 antibody. n = 3 independent biological replicates. n, o TwoMP analysis of purified AIF (100 nM, n) and AIF3 (100 nM, o) monomer and/or dimer. p Docking model of full-length mature AIF or AIF3 with NADPH. Green, FAD binding ___domain; Blue, NADH binding ___domain; Purple, C-terminal ___domain; Red, exon 1-encoded peptide; Cyan, peptide encoded by a portion of exon 2, located before the hydrophobic ___domain; Yellow, hydrophobic ___domain encoded by partial exons 2 and 3 inserting into the membrane; Brown, peptide encoded by the remaining exon 3 and the initial 9 aa in exon 4; White circle, NADPH binding pocket.
Previous studies showed that AIF residues, including G307, F309, L310, E313, E335, G398, S479, and H453, are involved in binding and stabilizing NADH19,37. Quadruple mutations of G307/F309/L310/E313 into alanine (QM-GFLE) significantly inhibited the NAD(P)H dehydrogenase activity in vitro (Fig. 5k and Supplementary Fig. 5g, h), while point mutations including E335A, G398A, S479A, and H453A failed to do so (Fig. 5k and Supplementary Fig. 5g, h). We also expressed and purified AIF protein with in-frame deletion of exon 2 (Δ2) or exon 3 (Δ3) from bacteria (Supplementary Fig. 5i). By comparison to WT AIF, the NADPH dehydrogenase activity of AIF-Δ2 was reduced by approximately 50% (Fig. 5l). In contrast, AIF-Δ3 had a full NADPH dehydrogenase activity comparable to WT AIF (Fig. 5l). These findings indicate that loss of exon 2, rather than exon 3, primarily contributes to the reduction of NAD(P)H dehydrogenase activity of AIF3.
To further understand whether loss of exons 2 and 3 in AIF3 causes confirmational changes contributing to the disruption of NAD+/NADH and NADP+/NADPH homeostasis, we expressed and purified both AIF and AIF3 proteins from bacteria in the absence of β-mercaptoethanol or dithiothreitol (DTT) in the native buffer system. AIF3 exhibited both monomeric and dimeric forms (Fig. 5m and Supplementary Fig. 5j), and had a higher average melting temperature (Tm) compared to AIF under different buffer or substrate conditions, including NaCl vs. MgCl2 buffer systems and NADH vs. NADPH substrates (Table S3). Moreover, purified AIF3, but not AIF, displayed a distinct dimer peak as determined by the Refeyn TwoMP Mass Photomertry system (Fig. 5n, o). These findings indicate that AIF3 has a tighter and more stable structure than WT AIF in vitro.
Although the crystal structure of short “soluble” AIF protein has been previously resolved, all these structures were notably generated from truncated AIF proteins lacking part or all of exons 2 and 3-encoded peptide, and thus they may not represent the mature AIF protein structure in nature. Using ColabFold and CB-Dock2 (https://cadd.labshare.cn/cb-dock2/php/index.php) programs, we built docking models of mature AIF/NADPH and AIF3/NADPH (Fig. 5p). Mature AIF and AIF3 proteins differed in the peptide encoded by exons 2 and 3, which constituted a hydrophobic ___domain (yellow in 1st left panel, Fig. 5p) inserted into the inner membrane. This hydrophobic ___domain (yellow) together with the peptide encoded by the remaining portion of exon 3 and the initial part of exon 4 (brown) locked AIF in the inner membrane and acted as a door holder to guide NAD(P)H access to the active binding sites (Fig. 5p, 1st left panel). However, AIF3 contained a free tail encoded by exon 1 (red in 2nd left panel, Fig. 5p), which may impede NAD(P)H’s access to the active binding site. Furthermore, dimeric AIF3 had a substantial structural change, which blocked NAD(P)H’s access to the active binding sites (Fig. 5p, right two panels). These findings indicate that the loss of exons 2 and 3-encoded peptide leads to a conformational change of AIF3 protein, which subsequently abolishes the NAD(P)H dehydrogenase activity and disrupts NAD(P)+/NAD(P)H homeostasis.
AIF3 splicing abolishes AIF’s GR activity to block GSH regeneration and ROS clearance
Three pathways, including catalase, thioredoxin system, and GSH system, are responsible for ROS clearance (Fig. 6a)38,39. To identify which pathway is dysregulated by AIF3 splicing, we examined protein levels of key enzymes involved in these three pathways. GPX4 was robustly decreased in AIF3 splicing MEFs, AIF3 splicing mouse brain, and AIF3 Tg mouse brain (Fig. 6b, quantification in Source Data). In contrast, PRDX1 had a negligible decrease, while GR, catalase, and MnSOD remained unchanged (Fig. 6b). We showed that overexpression of GR successfully cleared H2O2-induced ROS in AIF3 splicing MEFs (Fig. 4j). GR controls GSH regeneration, a process involving NADPH oxidation, leading us to hypothesize that AIF increases GSH regeneration contributing to ROS clearance (Fig. 6a). To test this, we examined whether the GSH pathway is dysregulated by AIF3 splicing. The metabolomic analysis showed that both GSH and GSSG levels were increased in AIF3 splicing MEFs, which could not be reversed by re-expression of AIF (Fig. 6c, d). However, the ratio of GSH/GSSG was significantly reduced, indicating that the reaction between GSH and GSSG favored the formation of GSSG, with limited GSH regeneration, in AIF3 splicing MEFs (Fig. 6e). Next, we studied whether AIF, AIF3, and AIF3L (starting at exon 4) regulate the GR activity by reducing GSSG to GSH in an in vitro biochemical assay. AIF, AIF3, and AIF3L proteins were expressed and purified from bacteria and their protein purity was verified by Coomassie blue staining and immunoblot assay (Supplementary Fig. 6a, b). AIF protein significantly enhanced the GR activity, whereas AIF3 or AIF3L protein failed to do so (Fig. 6f).
a Scheme of three main ROS clearance pathways and a proposed role of AIF in ROS clearance. b Effects of AIF3 on the expression of key enzymes in ROS clearance pathways. n = 3 biological replicates. c–e Metabolomics analysis of GSH/GSSG in AIF3 splicing MEFs. n = 5 biological replicates. f GR activity in the presence of 3 μM purified AIF, AIF3, or AIF3L. n = 17 (GR/GR + AIF/GR + AIF3), and 14 (GR + AIF3L) biological replicates. g Structural comparison of AIF (PDB 1gv4) and GR (PDB 1ger). C, C-terminal ___domain. h GR activity of 3 μM purified AIF and AIF3 in time-dependent manner. n = 3 (Auto/AIF3), and 6 (AIF) biological replicates. i GR activity of purified AIF, AIF3 (3 μM), and GR (0.1 mU). n = 7 (Auto), 6 (AIF), 3 (AIF3), and 4 (GR) biological replicates. j Expression of AIF/AIF3 and GR proteins in AIF3 splicing, AIF KO, GR KO, AIF-GR DKO, and DKO/AIF(3) rescue MEFs. k, l Effects of AIF and AIF3 on GR activity in cells established in (j). n = 6 biological replicates. m GR activity in the forebrain of WT and AIF3 splicing mice (Emx1-Cre + ). n = 12 (WT) and 16 (AIF3) biological replicates (unpaired 2-tailed Student’s t test). n GR activity of purified mouse AIF (mAIF) proteins and its mutants (3 μM). n = 7 biological replicates. o Analysis of mitochondrial ROS levels in AIF3 splicing, AIF KO and AIF-GR DKO MEFs treated with DMSO or H2O2 in the absence or presence of GSH. n = 9 (WT/AIF3) and 6 (AIFKO/DKO) biological replicates (two-way ANOVA with Sidak’s multiple comparison test). Data are shown as mean ± S.E.M throughout this figure. One-way ANOVA with Tukey’s multiple comparison test was applied in c–f, i, k, l, n.
Structural alignment analysis revealed that AIF (PDB:1GV4) exhibited a structure reminiscent of GR (PDB: 1GER), highly overlapped with the conserved NADH and FAD binding domains critical for the GR activity (Fig. 6g)19. Next, we asked whether AIF itself possesses the GR activity. Using the GSH colorimetric detection assay, we found that purified AIF protein (3 μM) demonstrated an evident GR activity in a time-dependent manner, whereas AIF3 protein exhibited only about 1/3 of AIF’s GR activity (Fig. 6h). These findings were further validated by GSH fluorometric detection assay, which showed AIF and AIF3 (3 μM) with GR activities of 43.8% and 18.5%, respectively (Fig. 6i). The activity of GR at 0.1 mU was set to be 100%. To further study the GR activity of AIF in intact cells, we generated GR knockout (GRKO) and GR/AIF double knockout (DKO) MEFs without or with sgRNA-resistant AIF or AIF3 expression (Fig. 6j). AIF3 splicing decreased about 25% of GR activity in MEFs, which could not be rescued by re-expression of AIF (Fig. 6k). Approximately a 40% reduction in GR activity was found in AIF KO MEFs, which is consistent with the in vitro GR activity data (Fig. 6i, k). Interestingly, about 30% of GR activity remained in GRKO MEFs (Fig. 6k), which was further decreased in DKO MEFs (Supplementary Fig. 6c, d). Overexpression of AIF, but not AIF3, in DKO MEFs restored GR activity to levels observed in GRKO MEFs (Fig. 6k, l). As expected, the GR activity was fully rescued by overexpression of E. coli GR in GRKO MEFs, GRKO AIF3 splicing MEFs, and DKO MEFs (Supplementary Fig. 6c, d). We further confirmed that the in vivo GR activity was decreased in the cortex of AIF3 splicing mice (Emx1-Cre + ) compared to littermate controls (Fig. 6m). GR protein expression was comparable in these mice (Fig. 6b). These findings demonstrate that AIF possesses a previously unrecognized GR activity and this activity is attenuated by AIF3 splicing.
Cysteine residues are critical for GR activity40. To determine the role of all three cysteine residues (C255, C316, and C440) located in the NADH and FAD binding domains of AIF in GR activity, we expressed and purified C255A, C316A, and C440A mutant AIF proteins from bacteria (Supplementary Fig. 6e). Both C255A and C440A AIF mutants exhibited reduced GR activity compared to WT AIF, whereas C316A mutant did not (Fig. 6n). None of these mutants (C255A, C316A, or C440A) affected the NAD(P)H dehydrogenase activity of AIF (Supplementary Fig. 6f, g). We further found that QM-GFLE mutation (G307/F309/L310/E313A), which blocks NADH binding to AIF, significantly decreased GR activity (Supplementary Fig. 6h). No GR activity was significantly reduced by individual AIF mutants, including E335A, G398A, S479A, and H453A (Supplementary Fig. 6h). These findings indicate that the GR activity of AIF is contingent on AIF protein structure and NAD(P)H/FAD binding.
Next, we assessed the effect of AIF3 splicing on thioredoxin reductase (TrxR) activity. AIF3 splicing caused a minor reduction in TrxR activity in MEFs (Supplementary Fig. 6i), indicating that the thioredoxin system is unlikely a major contributor regulating ROS clearance in AIF3 splicing cells. In contrast, in the presence of GSH, elevated mitochondrial ROS levels could be reduced in AIF3 splicing MEFs under oxidative stress conditions (Fig. 6o). However, GSH did not prevent the decline in mitochondrial membrane potential caused by AIF3 splicing (Supplementary Fig. 6j, k). Taken together, these findings suggest that AIF3 splicing primarily affects the GSH system, resulting in failure of ROS clearance.
Restoring NADH homeostasis by NDI1 reduces ROS levels and prevents neuronal loss in AIF3 splicing mouse brain
We next investigated whether restoring NADH homeostasis counteracts AIF3 splicing to prevent brain development deficits in mice. Yeast NADH dehydrogenase NDI1 acts as an alternative complex I (type 2 NADH-ubiquinone oxidoreductase) that catalyzes the transfer of an electron from NADH41. Thus, we tested whether NDI1 expression prevents AIF3-triggered mitochondrial dysfunction and ROS accumulation. We confirmed that purified NDI1 exhibited both NADH and NADPH dehydrogenase activities in vitro, similar to AIF, with relatively stronger activity for NADH (Supplementary Fig. 7a-c). Using UQ1, a derivative of the mitochondrial electron transport chain cofactor CoQ10, as the electron acceptor, we observed that both AIF and NDI1 exhibited comparable but significantly enhanced NADH oxidase activity compared to when O2 was the electron acceptor, whereas AIF3 significantly reduced NADH oxidase activity (Fig. 7a). NDI1 expression reversed an increase in NAD(P)H levels and decreases in NAD(P)+ levels as well as NAD(P)+/NAD(P)H ratio in AIF3 splicing MEFs (Fig. 7b–d and Supplementary Fig. 7d–f). The activity of mitochondrial complex I was also restored in AIF3 splicing MEFs when NDI1 was expressed (Fig. 7e). However, AIF did not phenocopy NDI1 to restore NADH and NADPH homeostasis and the complex I activity in AIF3 splicing MEFs (Fig. 7b–e and Supplementary Fig. 7d–f). We then performed intracerebroventricular (ICV) injection with adeno-associated virus (AAV) carrying NDI1 or GFP control into brains of AIF3 splicing mice (Emx1-Cre) and littermate controls at P0. In line with in vitro studies, NDI1 rescued NAD(P)+/NAD(P)H homeostasis in the cortex and hippocampus of AIF3 splicing mice at P21 (Fig. 7f–h and Supplementary Fig. 7g–i) and the mitochondrial complex I activity in the cortex of AIF3 splicing mice (Fig. 7i). The expression of NDI1 counteracted AIF3 splicing to effectively reduce cellular ROS levels in MEFs upon H2O2 treatment (Fig. 7j). However, NDI1 did not display GR activity with purified proteins in vitro (Supplementary Fig. 7j, k) nor alter GR activity in GRKO MEFs (Supplementary Fig. 7l, m). NDI1 expression also failed to alter total and mitochondrial ATP levels in AIF3 splicing MEFs (Supplementary Fig. 7n, o), which was expected as NDI1 does not contribute to proton pumping while transferring an electron. Studies from TMRE staining, AIF/MitoTracker staining, and mitochondrial complex subunit expression showed that NDI1 expression did not rescue mitochondrial membrane potential, mitochondrial morphology, and protein stability of complex subunits in AIF3 splicing MEFs (Supplementary Fig. 7p–s, quantification in Source Data). These findings indicate that NDI1 expression rescues mitochondrial complex I activity and reduces ROS levels in AIF3 splicing cells and brain by restoring cellular NAD(P)+/NAD(P)H homeostasis.
a NADH dehydrogenase activity of purified AIF, AIF3 and NDI1 with or without electron acceptor UQ1 (100 μM). Data are shown as mean, n = 3 biological replicates. b–d Effects of NDI1 on NADH/NAD+ levels in AIF3 splicing MEFs. n = 4 biological replicates. e Mitochondrial complex I activities in AIF3 splicing and rescue MEFs. n = 4 biological replicates. f–h NADH/NAD+ in the hippocampus of WT and AIF3 splicing (Emx1-Cre + ) mice injected with AAV-GFP or AAV-NDI1. The values of no injection-WT and -AIF3 splicing mice from Fig. 5h-j were used as controls. n = 10 biological replicates. i Mitochondrial complex I activities in the cortex of WT and AIF3 splicing (Emx1-Cre + ) mice injected with AAV-GFP or AAV-NDI1. n = 6 (WT-GFP/AIF3-GFP), 6 (WT-NDI) and 10 (AIF3-NDI) mice. j Mitochondrial ROS levels in AIF3 splicing and rescue MEFs treated with H2O2. n = 3 biological replicates. k, l Nissl staining and quantification of cortical thickness at the identical positions of AIF3 splicing mice (Emx1-cre + ) and their littermates injected with AAV-GFP or AAV-NDI1. n = 27 (WT), 16 (AIF3), 11 (WT-GFP), 14 (AIF3-GFP), 25 (WT-NDI1) and 33 (AIF3-NDI1) images from 4 (WT) and 5 (AIF3) mice/group. m–p Expression (m, n) and quantification (o, p) of CUX1 and NeuN in Emx1-Cre (m) or iCre (n)-induced AIF3 splicing mice and their littermates injected with AAV-GFP or AAV-NDI1. n = 6 mice/group (o) and 15 images/group (p) prepared from 4 WT/WT-NDI1 mice, 3 AIF3 splicing mice, and 6 AIF3-NDI1 mice. q, r NDI1 expression in Emx1-Cre (q) or iCre (r)-induced AIF3 splicing mice and their littermates injected with AAV-GFP or AAV-NDI1. n = 5 (WT) (q), 4 (WT) (r) and 6 (AIF3) (q, r) mice/group. Scale bar, 200 μm (k, m, n, q, r). Data are shown as mean ± S.E.M throughout this figure unless otherwise indicated. One-way ANOVA with Tukey’s multiple comparison test was applied in b-e, and Two-way ANOVA with Tukey’s multiple comparison test was applied in f–j, l, o, p.
Next, we studied whether NDI1 expression prevents AIF3-triggered neuronal cell death and mouse brain development deficits using two different AIF3 splicing mouse models, in which AIF3 splicing was induced embryonically (Emx1-Cre) or postnatally at P0 (CamkIIα-iCre). The expression of NDI1 or GFP was introduced via ICV injection in AIF3 splicing mice and their littermate controls at P0 (Fig. 7k-r). Nissl staining revealed a marginal increase in cortical thickness by NDI1 expression in Emx1-Cre-induced embryonic AIF3 splicing mice at P21 (Fig. 7k, l). These findings were confirmed by immunostaining of CUX1 and NeuN, demonstrating that NDI1 expression partially restored CUX1 expression in the cortex of AIF3 splicing mice (Fig. 7m, o). In contrast, NDI1 expression had no obvious rescued effect on hippocampal morphology in Emx1-Cre-induced AIF3 splicing mice (Fig. 7k). Notably, we observed a substantial rescued effect of NDI1 on the thickness of CUX1+ cortex layer in CamkIIα-iCre-induced postnatal AIF3 splicing mice at P60 (Fig. 7n, p). The expression of NDI1 in the cortex of mouse brain was verified by immunostaining (Fig. 7q, r). These findings indicate that NDI1 expression partially protects against AIF3 splicing-induced cortical neuron loss in mice during brain development.
Discussion
The major finding of this study is that AIF is a mammalian NAD(P)H dehydrogenase and possesses previously unrecognized GR activity, essential for maintaining cellular GSH-redox homeostasis and mitochondrial functions through dual regulatory mechanisms, including safeguarding mitochondrial integrity and OXPHOS to eliminate ROS generation as the first line of defense and facilitating GSH regeneration to support the antioxidant defense system for ROS clearance as the second line of defense. Our study has revealed that mutations and variants in exons 2 and 3 of AIFM1 are the risk factors to enhance the frequency of pathologic AIF3 splicing in brain of pediatric individuals with mitochondrial disorders. AIF3 splicing disrupts mitochondrial cristae structure, membrane potential, and OXPHOS, causing notable brain development deficits. Mechanistically, AIF3 lacks GR and NAD(P)H dehydrogenase activities, causing mitochondrial dysfunction and dysregulation of GSH-redox homeostasis. Expression of yeast NDI1 partially restores mitochondrial functions and protects cortical neurons in AIF3 splicing mice.
To date, in humans, AIF mutations have been identified at least at twenty-six different residues mainly located in the N-terminal oxidoreductase ___domain, which cause severe mitochondriopathies with neurodegeneration as a common feature1,2,3,4,5,6,7,8,9,10,11,12. In rodents, Harlequin mouse displays another type of naturally occurring AIF mutant with a proviral insertion at the intron 1 region, causing gene silencing and subsequent mitochondrial OXPHOS defect and progressive neurodegeneration13. Our study uncovered a remarkably high incidence of exon 2-deleted AIF (ΔE2AIF) variant in about 50% of normal human autopsy brain tissues and, notably, an even greater prevalence (100%) in pediatric individuals with mitochondrial disorders. ΔE2AIF is a splicing variant in addition to two currently known splicing options at exon 2a or 2b, indicating that exon 2 is a hotspot for splicing. Although ΔE2AIF is expected to cause a frame-shift, resulting in loss of AIF protein, those humans with a small portion of ΔE2AIF variant did not show obvious neurological abnormalities. Interestingly, the whole exome sequencing data analysis of 141, 456 humans demonstrated a significantly high genomic mutation rate in exon 3 of AIFM1. Genome editing at exons 2 and/or 3 using CRISPR/Cas9 technique directly revealed that genomic mutations/variants in exons 2 and 3 enhance AIF3 splicing, which provide a fundamental theoretical support for our recently established AIF3 splicing mouse models. This also explains why our original “conditional knockout strategy” by deleting exon 3 of Aifm1 using the Cre-LoxP system to knock out AIF fails36. Instead, an endogenous splicing machinery is activated to facilitate an “imperfect” AIF3 splicing ensuring cell survival. Importantly, our unbiased analysis of human AIF splicing variants further confirmed that AIF3 and AIF3L splicing isoforms occur at a relatively high frequency in all 7 postmortem brain tissues from pediatric individuals with mitochondrial disorders, highlighting a potential role of these aberrant AIF splicing isoforms in disease progression.
Given that AIF and AIF3 are encoded by the same AIFM1 gene, AIF3 splicing involves two events: a reduction in AIF and an increase in AIF3. Then, an important question would be whether loss-of-AIF and/or gain-of-AIF3 contribute to AIF3 splicing-triggered mitochondrial dysfunction and subsequent brain development deficits. Previous studies have shown that AIF KO mice are embryonically lethal15. Mice with AIF3 splicing induced during the embryonic stages by Nestin-cre or Emx1-Cre survive at birth with brain development deficits, indicating that AIF3 splicing does not simply phenocopy AIF KO. It might preserve partial AIF functions and gain additional new functions. The latter is supported by the fact that AIF3 overexpression increases the density of mitochondrial cristae and reduces the OCR and activities of the complex IV and V. Although AIF3 splicing shares many common features with AIF loss including reduced mitochondrial complex stability and OXPHOS, our studies from MEFs showed that AIF3 splicing exhibits much worse effects on mitochondrial dysfunction than AIF3 overexpression or AIF KO. It is worth pointing out that AIF KO MEFs we used here are a mosaic and genetically heterogeneous population resulting from sgRNA-mediated genome editing and clonal selection. AIF KO is expected in most, but not all, MEFs. For all experiments, we used the first five passages of AIF KO MEFs after clonal selection with puromycin, and AIF KO was confirmed by western blot. Thus, AIF KO MEFs are better regarded as “AIF knockdown” MEFs, which might mimic Harlequin mice with less severe detrimental phenotypes as observed in genetic deletion of AIF cells or mice. Therefore, our studies, together with others, demonstrate that the effects of AIF loss and AIF3 splicing on mitochondrial dysfunction rank in the following order: genetic deletion of AIF > AIF3 splicing > AIF knockdown > AIF3 overexpression.
Although AIF is a ubiquitous oxidoreductase with conserved FAD- and NADH-binding domains overlapping with the GR ___domain from a structural perspective, so far, no GR or NADPH dehydrogenase activity has been reported for AIF protein to our knowledge. Previous studies showed that AIF has no or much weaker NADH oxidase activity than its yeast homolog NDI123. Thus, AIF has not been typically considered as an alternative NAD(P)H dehydratase in humans as NDI1 does in yeast. Our studies clearly demonstrated that AIF exhibits NADH dehydrogenase activity comparable to NDI1 and efficiently oxidizes NADH to NAD by transferring electrons to ubiquinone rather than oxygen. Moreover, AIF even possesses previously unrecognized NADPH dehydrogenase and GR activities, which are not identified for NDI1. The discrepancy between our findings and others is likely due to the fact that full-length mature AIF is purified and used in our study for its enzymatic analyses, whereas other research studies truncated AIF protein, which lacks the part or all of exons 2 and 3-encoded peptide. Our results provided direct evidence that the exons 2-3-encoded hydrophobic ___domain is another motif critical for its dehydrogenase activity, in addition to FAD/NAD(P)H binding pocket. In line with this, AIF3 displays reduced NAD(P)H dehydrogenase and GR activities as shown in our current study. Molecular docking modeling of AIF(3) and in vitro biochemical assays suggest that AIF3 exhibits greater solubility and stability to form a much tighter dimer, which may create steric hindrance within the narrow catalytic pocket and likely contributes to its reduced enzymatic properties compared to AIF protein. In contrast, AIF contains an insoluble N-terminal transmembrane peptide, which serves as a door holder opening a path and guiding NADH and/or NADPH to access the catalytic pocket. Thus, this peptide is critical for NAD(P)H dehydrogenase and GR activities of AIF. Previous studies showed that Cys58 and Cys63 are two key residues on human GR protein that form a disulfide bond critical for GR activity40. Interestingly, our current study found that, besides key residues in the NAD(P)H binding pocket, Cys255 and Cys440 are essential for AIF’s GR activity. However, Cys255 and Cys440 are positioned slightly apart, which is less likely to directly form a disulfide bond, suggesting that these two Cys residues may undergo certain modifications such as CoAlation that facilitate the formation of a disulfide bond.
Although AIF itself is not part of the mitochondrial complex, our study demonstrated that AIF is essential for mitochondrial OXPHOS and cell survival via dual regulatory mechanisms. Under physiological conditions, electron flow through mitochondrial complexes is well-controlled, ultimately reducing molecular oxygen to water. However, mitochondrial complex I deficits often increase electron leakage, leading to reactions with molecular oxygen and elevated production of ROS42. AIF functions as an alternative NAD(P)H dehydrogenase when mitochondrial complex I has functional deficits, since it has the ability to efficiently transfer electrons from NADH to ubiquinone rather than O2 as we showed. Thus, AIF not only safeguards the integrity of mitochondrial complexes but also serves as a contingency, stepping in to fulfill crucial functions in OXPHOS during times of compromise, acting as the first line of defense against ROS generation. In contrast, AIF3 splicing not only causes disassembly of mitochondrial complex I, III and IV but also fails to act as an alternative NAD(P)H dehydrogenase to bypass the complex I. Therefore, AIF3 splicing increases ROS generation as we showed here. GSH is an important antioxidant that scavenges ROS43. During this process, GSH is oxidized to form GSSG and GR helps regenerate GSH, which is crucial for maintaining an adequate pool of GSH. Interestingly, our findings demonstrated that AIF itself has GR activity and also cooperates with GR protein to enhance GR activity, thereby facilitating GSH regeneration for ROS clearance as the second line of defense. Unlike AIF, AIF3 lacks NADPH dehydrogenase and GR activities, thereby disrupting GSH-redox homeostasis. These dual regulatory mechanisms explain the importance of AIF physiological functions critical for life.
Our findings revealed that a single yeast enzyme NDI1 counteracts AIF3 splicing to partially restore mitochondrial complex I activity and to reduce ROS levels, thereby protecting cortical neurons in AIF3 splicing mice. However, NDI1 does not contribute to the maintenance of mitochondrial morphology, ATP production, and mitochondrial membrane potential. Restoring NADH dehydrogenase activity alone through NDI1 expression seems sufficient to replace mitochondrial complex I activity, but it does not fully restore mitochondrial functions, which likely require ATP production. Surprisingly, expression of AIF in AIF3 splicing cells has limited rescued effects on mitochondrial morphology, complex stability and other functions, suggesting that AIF3 might act as a dominant-negative competitor for AIF.
Limitations of the study
There are certain limitations in our work that should be further explored in future. First, while our findings reveal that both loss-of-AIF and gain-of-AIF3 new functions contribute to AIF3 splicing-triggered mitochondrial dysfunction and neurodegeneration, the precise functions AIF3 has lost or gained remain unknown. AIF plays an important role in metabolic regulation. In future, it is important to dissect the effects of AIF vs. AIF3 on metabolic changes. Second, it is important to study the molecular mechanism by which AIF maintains mitochondrial complex stability and assembly. Third, although our study highlighted the effect and underlying mechanism of AIF3 splicing-triggered mitochondrial disfunction, it remains unknown whether mitochondrial dysfunction alone is essential and sufficient to cause neurodegeneration and neuronal cell death. Given the fact that AIF(3) contributes to both death and life, the relationship between AIF3 splicing-triggered mitochondrial dysfunction and neurodegeneration in aging should be further explored. Lastly, uncovering the detailed AIF3 splicing mechanism would help better develop therapeutic interventions protecting brain from aberrant AIF splicing variants-induced mitochondrial dysfunction and neurodegeneration.
Methods
Ethical statement and human postmortem brain tissue collection
All experiments conducted in this study comply with relevant ethical regulations. Human samples used in this study were de-identified and collected from deceased subjects. The UT Southwestern Institutional Review Board (IRB) has classified research involving “deceased subjects” as “Non-Human Research” and exempt from human subject regulations as codified in federal law. Snap-frozen frontal and/or temporal (including hippocampal) autopsy brain tissue samples from age and gender-matched cases diagnosed with or without mitochondrial disorders were obtained from the Children’s Medical Center Pediatric Biospecimen Repository and the William P. Clements Jr. University Hospital at UT Southwestern Medical Center. The autopsies were performed within a timeframe of 12 to 72 h after death and the samples were frozen at a temperature of −80 °C. The tissues were sourced with consent for research purposes obtained for all autopsy samples and were deidentified. No individually identifiable protected health information was accessible to the investigators using these materials. Their essential information including sex and age range can be found in Tables S1 and S2.
Animals
A tissue-specific conditional AIF3 splicing mouse model was established via conditional deletion of exon 3 of Aifm1 using Cre-LoxP system, and an AIF3 transgenic mouse model was generated by inserting LoxP-eGFP/STOP-LoxP-AIF3 into Rosa26 locus, as described in our recent study36. The CamkIIα-iCre mouse line (EM:01153) was obtained from EMMA. Emx1-Cre and Nestin-Cre mice were from Jax (Cat #005628 and #003771). All mice were housed and cared at UT Southwestern Medical Center animal facility with a 12-h light/dark cycle in accordance with the guidelines outlined in the NIH Health Guide for the Care and Use of Laboratory Animals. UT Southwestern Medical Center is fully accredited by the American Association for the Accreditation of Laboratory Animal Care (AAALAC), ensuring compliance with animal welfare standards. The research procedures conducted in this study were approved by the Medical Institutions Institutional Animal Care and Use Committee (IACUC), adhering to the Animal Welfare Act Regulations and Public Health Service (PHS) policy.
Cell culture
WT and AIF3 splicing MEFs were prepared from AIFfl/fl mice and infected with or without Cre lentivirus in DMEM with 10% heat-inactivated fetal bovine serum at 37 °C in a 5% CO2/95% air incubator as described previously36. All KO cell lines were generated using the CRISPR/Cas9 technique and genotyped as described44. All cell lines were mycoplasma-free. AIF and AIF3 primary neuronal cultures were prepared from the cortex as described previously36. Neurons were maintained in MEM with 10% horse serum, 30 mM glucose, and 2 mM L-glutamine in a 7% CO2 humidified 37 °C incubator. The growth medium was refreshed three times per week.
Plasmid constructs
Full-length mouse AIF or AIF3 cDNA was amplified by PCR and cloned into cFUGW-3xFlag-C vector as described previously36. Full-length human AIF and truncated AIF cDNA was amplified by PCR and cloned into pGEX-6p-1 vector. Full-length E.coli GR cDNA was amplified by PCR and cloned into pLVX-Ubc-FLAG vector. Full-length SaNDI1 was subcloned into pLVX-Ubc-FLAG and pGEX-6p-1 vectors from NDI1 plasmid (Addgene, #72876). sgRNA-resistant full-length mouse AIF or AIF3 cDNA was amplified by PCR and cloned into pLVX-Ubc-FLAG vector.
Lentivirus production
Lentivirus production was performed as described previously36,45. In brief, 5.4 µg transducing vector, 6 µg psPAX2, and 1.8 µg pMD2.G using 13.2 µL of polyethyleneimine (Fisher, NC1038561, 5 mg/mL) were transiently transfected to HEK293T cells. Lentivirus was harvested at 48 h and 72 h after transfection and concentrated by ultracentrifugation for 1 h at 100,000 g at 4 oC.
AAV production
AAV production was performed as described previously46. In brief, pAAV-CBH-2HA (Wang Lab), pUCmini-iCAP-PHP (Addgene, #103005), pAd-deltaF6 (Addgene #112867) plasmids were co-transfected into HEK293T cells at the ratio of 1:4:2 using polyethyleneimine (Fisher, NC1038561, 5 mg/mL). Three to five days after transfection, viruses were harvested from both cell culture media and cells. The harvested viruses were first centrifuged at 2000 g for 15 minutes at room temperature. The resulting pellet was resuspended in 5 mL of salt-active nuclease (SAN, 25 U/μL; ArcticZymes, cat. # 70910-202) at 37 °C for 1 h and stored at 4 °C (AAV collection #1). In parallel, the resulting supernatant (5 volumes) was treated with 40% PEG stock solution (1 volume) on ice for 2 h or 4 °C overnight. After centrifugation at 4000 g for 30 minutes at 4 °C, the pellet was resuspended in 5 mL of SAN at 37 °C for 1 h as AAV collection #2, which was eventually combined with AAV collection #1 and stored at 4 °C. The AAV virus was then purified using a 15%, 25%, 40%, and 60% iodixanol density gradient ultra-centrifugation at 350,000 g for 4 h. The virus was collected from the 40/60% interface and 40% layer. The collected AAV virus was further processed by dialyzing three times with DPBS and subsequently concentrating to the desired titer. The viral titers were determined by qPCR using ITR primers (Forward, 5’-GGAACCCCTAGTGATGGAGTT-3’; reverse, 5’-CGGCCTCAGTGAGCGA-3’).
RNA in situ hybridization assay
RNA in situ hybridization assay was performed on paraffin embedded cortex/hippocampal slices from control and pediatric patients with mitochondrial disorders. 5 μm sections were cut, deparaffinized and rehydrated by immersing the slides through Xylene and Ethanol. Slides were then washed twice in DEPC-PBS, followed by digestion with 10 μg/mL Proteinase K for 30 min at room temperature. Afterward, the slides were washed once in DEPC-PBS at room temperature for 5 min, fixed in 4% PFA in DEPC-PBS for 15 min, and rinsed twice in DEPC-Water. After removing the DEPC-water and adding 15 mL Triethanolamine-acetic anhydride solution for 10 min at RT, slides were washed in DEPC-PBS at room temperature for 5 min and pre-hybridize in hybridization solution (50% Formamide, 5 × SSC, 0.3 mg/mL Yeast tRNA, 100 μg/mL Heparin, 1 × Denhardt’s Solution, 0.1% Tween 20, 0.1% CHAPS, 5 mM EDTA) for 1-4 h in a 65 °C water bath. Next, the slides were incubated with the specific probe in the hybridization buffer for 12-16 h or more. After three times washing with 2 × SSC for 15 min at 65 °C, the slides were then incubated with 1.0 μg/mL RNase A in 2 × SSC at 37 °C for 30 min. The sides were further proceeded with two washing steps, including three times washing with 0.2 × SSC for 15 min at 65 °C and 2 times washing in PBST (0.1% Triton X-100 and 2 mg/mL BSA in PBS) for 10 min each at room temperature. Subsequently, the slides were blocked in PBT + 10% heat-inactivated sheep serum for 30 min to 1 h at room temperature and incubated with anti-DIG antibody overnight at 4 °C. Finally, after washing 3 times in PBT for 30 min each at room temperature and two times in alkaline-phosphatase (AP) buffer (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 0.1% Tween20, 50 mM MgCl2) for 5 min each at room temperature, the slides were incubated with 1 μL 75 mg/mL NBT and 3.5 μL 50 mg/mL BCIP per mL AP buffer and developed in the dark for 2-20 h. The images were visualized using Nikon bright-field microscopy (ECLIPSE Ci, M568E).
5’ RACE assay
5′-RACE assay (Qiagen) was performed using the cDNAs prepared from postmortem brain tissues of pediatric patients with mitochondrial disorders as described previously36. The first round of PCR amplification was performed using the adaptor primer UPM (5’-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3’) and AIFM1-specific reverse primer GSP1 (5’-GCCCCACAGTGGAATTGGCAAACCCACT-3’). The nested PCR was performed with the universal short primer (5’-CTAATACGACTCACTATAGGGC-3’) and AIFM1 -specific reverse primer NGSP1 (5’-GGTGGGGGCTTCCTTGGTAGCGAACTGG-3’). PCR products were then cloned using In-Fusion HD cloning kit (TAKARA) and sequenced.
Immunostaining assay
Immunostaining assay was performed as described previously36,44. For immunocytochemistry, cells were fixed with 4% paraformaldehyde, permeabilized with 0.01% Triton X-100, and blocked with 3% BSA in PBS. For immunohistochemistry, brain sections (30 μm) were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 10% normal donkey serum in PBS. Brain sections/cells were then incubated with the specific primary antibody followed by fluorescent secondary antibody as indicated in Key Resources Table. Immunofluorescent and immunohistochemical analysis were carried out by using a fully motorized Zeiss Observer Z1 fluorescence microscope (Carl Zeiss).
Immunoblot and immunoprecipitation (IP) assays
Immunoblot assay was performed as described previously44. Cells were lysed in modified RIPA buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1 mM β-mercaptoethanol, 1% Igepal, 0.5 % sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonylfluoride (PMSF), 1 mM Na3VO4, 1 mM NaF, and fresh protease inhibitor cocktail). Equal amount of proteins were separated by SDS-PAGE gel and transferred to nitrocellulose membrane. The membrane was blocked with 5% milk and incubated overnight with primary antibodies as listed in Key Resources Table. For IP, cells were lysed in NETN lysis buffer (150 mM NaCl, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0, 0.5% Igepal, and fresh protease inhibitor cocktail) as described previously47. The supernatant was incubated with AIF-E1 (Santa Cruz, sc13116) or immunoglobulin G antibody overnight in the presence of protein A/G magnetic beads (Bio-Rad) at 4 °C. Precipitated proteins were separated by SDS-PAGE gel, followed by immunoblot assay.
Transmission electron microscopy
Cells were fixed on MatTek dishes with 2.5% (v/v) glutaraldehyde in 0.1 M sodium cacodylate buffer. After five rinses in 0.1 M sodium cacodylate buffer, they were post-fixed in 1% osmium tetroxide plus 0.8% K3[Fe(CN6)] in 0.1 M sodium cacodylate buffer for 1 h at room temperature. Cells were rinsed with water and en bloc stained with 2% aqueous uranyl acetate for 1 h. After five rinses with water, specimens were dehydrated with increasing concentrations of ethanol, infiltrated with Embed-812 resin, and polymerized overnight in a 60 oC oven. Epoxy discs were removed by submerging MatTek dishes in liquid nitrogen. Blocks were sectioned with a diamond knife (Diatome) on a Leica Ultracut UCT ultramicrotome (Leica Microsystems) and collected onto copper grids, post stained with 2% Uranyl acetate in water and lead citrate. Images were acquired on a JEOL 1400+ transmission electron microscope (FEI) equipped with a LaB6 source using a voltage of 120 kV and an AMT-Biosprint 16 M CCD camera system.
Mice were perfused through the heart with PBS (pH 7.4) for 30 s, and then perfused with 4% paraformaldehyde, 1% glutaraldehyde, 0.1 M sodium cacodylate buffer. Mouse cortical and hippocampal tissues were minced into 1 mm as described36 and fixed with 2.5% (v/v) glutaraldehyde in 0.1 M sodium cacodylate buffer. Tissue samples were then rinsed in 0.1 M sodium cacodylate buffer and post-fixed in 1% osmium tetroxide and 0.8% potassium ferricyanide in 0.1 M sodium cacodylate buffer for one and a half h at room temperature. After three rinses in water, tissues were en bloc stained with 4% uranyl acetate in 50% ethanol for two h. Next, tissues were dehydrated with increasing concentrations of ethanol, transitioned into resin with propylene oxide, infiltrated with Embed-812 resin, and polymerized overnight in a 60 °C oven. Blocks were sectioned with a diamond knife (Diatome) on a Leica Ultracut UCT ultramicrotome (Leica Microsystem) and collected onto copper grids, post-stained with 2% aqueous uranyl acetate and lead citrate. Images were acquired with a Tecnai G2 spirit transmission electron microscope (FEI, Hillsboro, OR) equipped with a LaB6 source at 120 kV using a Gatan Ultrascan CCD camera.
Seahorse assay
The OCR was measured in a Seahorse XF24e extracellular flux analyzer as described previously48. Briefly, cells were seeded at 0.5 × 105/well. Oligomycin (1 μM), FCCP (1 μM), antimycin A (5 μM), and rotenone (1 μM) were added into 4 different ports of the Seahorse cartridge, respectively. OCR was measured with a standard 8-min cycling program including mixture (3 min), waiting (2 min), and measurement (3 min). After Seahorse analysis, cells were lysed in 0.1 M NaOH and cellular protein concentrations were determined by Bradford assay for data normalization.
Mitochondrial DNA mass assay
Mitochondrial DNA mass was quantified as described previously36. Briefly, the relative abundance of mitochondrial DNA compared to nuclear genomic DNA was assessed using quantitative real-time PCR. This analysis involved the utilization of primers specific to the mitochondrial DNA-encoded cytochrome b gene and the nuclear DNA-encoded ribosomal protein S2 (RPS2) gene.
Subcellular fractionation assay
Freshly harvested brain cortex, hippocampus, or cells were washed once with ice-cold PBS, suspended in isolation buffer, consisting of 250 mmol/L sucrose, 10 mmol/L KCl, 1.5 mmol/L MgCl2, 1 mmol/L Na2EDTA, 1 mmol/L DTT, 1 mmol/L PMSF, 10 mmol/L Tris-HCl (pH 7.4), and protease inhibitor cocktail, and incubated on ice for 30 minutes, followed by homogenization. The homogenized cells/tissues were centrifuged at 100 × g for 5 minutes at 4 °C to remove unfractionated cells. The resulting pellet was the nuclear fraction that underwent two additional centrifugation steps at 700 × g for 10 minutes at 4 °C. The supernatant was then centrifuged at 12,000 × g for 15 minutes at 4 °C, resulting in the supernatant as the cytoplasm fraction and the pellet containing the mitochondrial fraction. The mitochondrial pellet was washed once with isolation buffer and resuspended in lysis buffer consisting of 30 mmol/L Tris-HCl (pH 7.4), 200 mmol/L KCl, 5 mmol/L EDTA, 0.5% Triton X-100, 1 mmol/L PMSF, and protease inhibitor cocktail. To prepare mitochondrial extracts, the resuspended mitochondria were subjected to three cycles of freezing in liquid nitrogen and thawing. The integrity of the mitochondria, cytoplasm and nuclear subcellular fractions was determined by monitoring CytC, α-tubulin and histone H3, respectively.
Submitochondrial fractionation assay
Submitochondrial fractionation was performed following mitochondrial isolation49,50. Briefly, the mitochondrial pellet was resuspended in lysis buffer consisting of 10 mmol/L KH2PO4 (pH 7.4), 1 mmol/L PMSF, and protease inhibitor cocktail, and the mixture was shaken at 4 °C for 15 min. An equal volume of buffer composed of 32% sucrose, 30% glycerol, 10 mmol/L MgCl2, 10 mmol/L KH2PO4 (pH 7.4), 1 mmol/L PMSF, and protease inhibitor cocktail was added to the mixture and shaken for another 15 min. The mixture was then centrifuged at 12,000 g for 10 min. The supernatant S1 and pellet P1 were collected. The pellet P1 was resuspended in 2 mL lysis buffer composed of 10 mmol/L KH2PO4 (pH 7.4), 1 mmol/L PMSF, and protease inhibitor cocktail, shaken at 4 °C for 30 min, and subjected to ultracentrifugation at 160,000 g for 60 min. The supernatant S2 (mitochondrial matrix) and pellet P2 (mitochondrial inner membrane) were collected. The supernatant S1 was also subjected to ultracentrifugation at 160,000 g for 60 min. The resulting supernatant (S3) was mitochondrial intermembrane space, and the pellet P3 was mitochondrial outer membrane.
Mitochondrial complex activity assay
The mitochondrial fraction was isolated as described above in “Subcellular fractionation” for the following complex activity assay. The activity of mitochondrial complex I was determined by measuring the oxidation of NADH with ubiquinone as the electron acceptor. Mitochondrial extracts (20 μg) were combined with prewarmed assay buffer (25 mmol/L KH2PO4, 25 mmol/L K2HPO4, pH 7.2, 2.5 mg/mL BSA, 5 mmol/L MgCl2, 2 mmol/L KCN, 0.01 mg/mL antimycin A, 1.3 mmol/L NADH, 0.01 mmol/L decylubiquinone) and incubated at 30 °C for 5 minutes. The absorbance at 340 nm was recorded every 30 seconds for 10 minutes at 30 °C using the Spark 10 M plate reader (TECAN), once a stable baseline was achieved. To stop the reaction, 0.25 mmol/L rotenone (10 μL) was added, and the absorbance at 340 nm was further monitored for 5 minutes.
The activity of mitochondrial complex II was assessed by monitoring the decrease in absorbance resulting from the oxidation of DCPIP at 600 nm. Mitochondrial extracts (20 μg) were diluted in lysis buffer. Simultaneously, assay buffer A and assay buffer B were prepared. Assay buffer A consisted of 25 mmol/L KH2PO4, 25 mmol/L K2HPO4 (pH 7.2), 20 mmol/L sodium succinate. Assay buffer B contained 25 mmol/L KH2PO4, 25 mmol/L K2HPO4, (pH 7.2), 2.5 mmol/L rotenone in ethanol, 2 mmol/L KCN, 0.01 mg/mL antimycin A, 50 μmol/L DCPIP. Both assay buffers A and B were prewarmed at 30 °C for 5 minutes. Just before initiating the reaction, the diluted mitochondrial extracts were mixed with a combination of assay buffers A and B. The absorbance at 600 nm was recorded every 30 seconds for 1 minute at 30 °C using a plate reader to establish a baseline. Subsequently, the absorbance at 600 nm was measured for an additional 10 minutes after adding 0.01 mmol/L decylubiquinone. While deficiencies in complex II are rare, several cases have been reported51,52. Complex II exhibits a rapid loss of activity following death and during improper storage. Consequently, its activity serves as an indicator of tissue integrity and the reliability of mitochondrial complex enzymology53.
The activity of mitochondrial complex III was determined by monitoring the increase in absorbance resulting from the reduction of cytochrome c at 550 nm. The assay buffer used for this measurement contained 50 mmol/L KH2PO4, 50 mmol/L K2HPO4 (pH 7.2), 20 mmol/L MgCl2, 0.1% BSA, 2.5 mmol/L rotenone, 1 mmol/L n-Dodecyl β-D-maltoside, and 2 mmol/L KCN. The assay buffer was prewarmed at 30 °C for 5 minutes. Freshly prepared 100 μmol/L DB·H2 and oxidized 25 μmol/L cytochrome c were used in the reaction. To establish a baseline, the absorbance at 550 nm was recorded every 30 seconds for 1 minute at 30 °C after adding DB·H2 and cytochrome c to the prewarmed assay buffer. The absorbance at 550 nm was then recorded for an additional 10 minutes after the addition of mitochondrial extracts (10 μg). As the reaction progressed, the color of the sample changed rapidly from orange to pink. At the end of the time course, 0.01 mg/mL antimycin A was added, and the absorbance at 550 nm was recorded for another 5 minutes.
The activity of mitochondrial complex IV was assessed by monitoring the decrease in absorbance resulting from the oxidation of cytochrome c at 550 nm. The assay buffer consisted of 20 mmol/L KH2PO4, 20 mmol/L K2HPO4 (pH 7.0), and 0.45 mmol/L n-Dodecyl β-D-maltoside. A freshly prepared solution of reduced cytochrome c at a concentration of 15 μmol/L was used. To establish a baseline, the absorbance at 550 nm was recorded every 30 seconds for 1 minute at 30 °C after prewarming the assay buffer at 30 °C for 5 minutes. Subsequently, the absorbance was recorded for an additional 10 minutes after adding mitochondrial extracts (10 μg). As the reaction progressed, the absorbance declined exponentially, and the color of the sample changed from pink to orange. At the end of the reaction, 2 mmol/L KCN was added, and the absorbance was recorded for 5 minutes.
The activity of mitochondrial complex V was determined by measuring the NADH oxidation through a two-step reaction that converts phosphoenolpyruvate to lactate. Mitochondrial extracts (20 μg) were mixed with 2.5 mmol/L ATP in prewarmed assay buffer (40 mmol/L Tris-HCO3, 2.5 mol/L phosphoenolpyruvate, 0.2 mmol/L NADH, 0.025 mg/mL antimycin A, 50 mmol/L MgCl2, 0.5 mg/mL lactate dehydrogenase, 0.1 mg/mL pyruvate kinase) and incubated at 30 °C for 5 minutes. The absorbance at 340 nm was recorded every 30 seconds for 10 minutes at 30 °C using a plate reader once a stable baseline was achieved.
Mitochondrial complex I, II, III, IV and V activities were normalized to citrate synthase activity48,54. To determine citrate synthase activity, the rate of coenzyme A (CoA.SH) production from oxaloacetate was measured by quantifying the levels of free sulfhydryl groups using the thiol reagent DTNB. DTNB readily reacted with sulfhydryl groups, resulting in the generation of free 5-thio-2-nitrobenzoate anions, which exhibited a yellow color that could be monitored at 412 nm. The assay buffer without oxaloacetate was prepared by combining the following components: 100 mmol/L Tris-HCl (pH 8.0), 100 μmol/L DTNB, 50 μmol/L acetyl coenzyme A, and 0.1% Triton X-100. It was then pre-warmed at 30 °C for 5 minutes before use. The baseline absorbance was recorded at 412 nm for 1 minute at 30 °C by adding 250 μmol/L oxaloacetate. After adding mitochondrial extracts (10 μg), the absorbance was recorded at 412 nm every 30 seconds for a total duration of 5 minutes at 30 °C.
BN-PAGE staining and in gel activity assays
BN-PAGE was used to separate mitochondrial protein complexes by NativePAGE Novex Bis-Tris Gel System (ThermoFisher Scientific) following the manufacturer’s instructions. Briefly, cell pellets were washed with cold PBS twice. Mitochondrial fractions were isolated as described above in “Subcellular fractionation” and then mixed with 5% digitonin solution in 1x native sample buffer (final concentration is 1%) on ice for 15 min. After centrifugation at 20, 000 g for 30 min at 4 oC, NativePAGE 5% G-250 Sample Additive was added right before loading samples onto the gel. Gels were run at 4 °C in a precooled apparatus with precooled samples and buffers. The 3% to 12% polyacrylamide gradient, as well as the buffers used for electrophoresis, was prepared as manufacturer’s instructions. After running, the gel was first incubated in ice-cold water for 15 min and then fixed with 100 mL of fix solution (40% methanol and 10% acetic acid) by heating in a microwave (950–1100 watts) for 45 seconds and shaking on an orbital shaker for 30 minutes at room temperature. Subsequently, the gel was incubated with 100 mL of Coomassie R-250 Stain (consisting of 0.02% Coomassie R-250 in 30% methanol and 10% acetic acid), microwaved (950–1100 watts) for 45 seconds, and then shaked on an orbital shaker for an additional 30 minutes. Finally, the gel was detained in 100 mL of destain solution (containing 8% acetic acid) by heating in the microwave (950–1100 watts) for 45 seconds and shaking on an orbital shaker at room temperature until the desired background was achieved.
For in-gel assessments, the samples and gels were prepared as described above and the resulting gels were transferred to a container and immersed in ice-cold water for 15 minutes. Subsequently, the gels were incubated in various complex substrate solutions for in-gel activity determination as follows. To evaluate Complex I activity, the gels were exposed to the solution containing 0.1 mg/mL NADH, 2 mM Tris/HCl, and 2.5 mg/mL Nitro Blue tetrazolium chloride at pH 7.4. For Complex II activity assessment, the gels were incubated in the medium containing 4.5 mM EDTA, 10 mM KCN, 0.2 mM phenazine methosulfate, 84 mM sodium succinate, 10 mM Nitro Blue tetrazolium chloride, in 1.5 mM phosphate buffer at pH 7.4. To achieve Complex IV staining, the gels were incubated in 9 mL of 50 mM phosphate buffer (pH 7.4) containing 5 mg of 3,3′-diaminobenzidine, 1 mL of catalase (20 μg/mL), 10 mg of cytochrome c, and 750 mg of sucrose. To assess Complex V activity, the gels were incubated in the solution composed of 34 mM Tris, 270 mM glycine, 14 mM MgSO4, 0.2% Pb(NO3)2, and 8 mM ATP, at pH 7.855. The images were obtained using ChemiDoc imaging system (Bio-Rad).
ATP assay
ATP assay was performed using an ATP determination kit (catalog # A22066, Thermo Fisher Scientific) as described previously48. Briefly, 1 × 105 cells were seeded onto a 12-well plate and cultured. Cells were lysed by adding 500 μL of 1% Triton X-100 and subjected to measurement in Spark 10 M plate reader (Tecan) and the luminescence was read according to manufacturer’s instructions. 5 mM 2-DG was pretreated to inhibit glycolysis for 2 h before harvesting cells in the presence of 10 mM glucose. The ATP levels were normalized to cellular protein concentrations quantified by Bradford assay.
ROS assay
Cells (1 × 104 cells/well) were seeded onto a 96-well plate and cultured. On the next day, cells were treated with either 250 μM H2O2 or DMSO for 0-2 h. Cells without treatment were used as controls. 5 mM GSH was used to clear ROS in the presence or absence of 250 μM H2O2. After the treatment, the medium was changed, and cells were allowed to recover for 0-6 h. Subsequently, cells were incubated with 5 μM MitoSOX for 10 minutes at 37 °C, trypsinized, and resuspended in 0.2 mL PBS. The total fluorescence representing ROS levels was measured using Tecan Spark 10 M plate reader, following the manufacturer’s instructions. The ROS levels were normalized to cellular protein concentrations quantified by Bradford assay.
Mitochondrial membrane potential assay
Mitochondrial membrane potential was assessed with 50 nM TMRE as previously described48. FCCP (10 μM) was used 30 min prior to TMRE staining as a positive control of mitochondrial membrane potential loss.
RNA-seq assay
RNA-seq was performed as described previously44. Total RNA was isolated from WT, AIF3 splicing and AIF3-AIF (rescue) MEFs using the RNeasy mini kit (Qiagen) and treated with DNase (Qiagen). The quality of total RNA was confirmed with a RNA integrity number score of 8.5 or higher by the Agilent TapeStation 4200. Library was prepared with the KAPA mRNA HyperPrep Kit (Roche) and sequenced on the Illumina NextSeq 2000 with the read configuration as 76-base pair (bp) single end. Bioinformatics were performed as described previously44.
Protein expression and purification
AIF (Δ53 aa), AIF3 (ΔE2–3), and other mutant AIF cDNAs were cloned into a pGex-6P-1 vector (GE HealthCare) at EcoR I and Xho I restriction sites. All AIF constructs were confirmed through Sanger sequencing. The proteins were then expressed and purified from E.coli C41(DE3) using GSH Sepharose36,56. Subsequently, the GST tag was removed using precision protease. The purity and concentrations of proteins were assessed by running a Coomassie blue gel, with bovine serum albumin as a reference.
NAD+/NADH assay
NAD+ and NADH levels were measured using NAD/NADH-Glo assay (G9071, Promega) as described previously48. Cells were lysed by adding 200 μL/well of 0.1 mol/L NaOH with 0.5% DTAB and subjected to measurement in Tecan Spark 10 M plate reader according to manufacturer’s instructions. The NAD+ and NADH levels were normalized to cellular protein concentrations quantified by Bradford assay (Bio-Rad).
NADP+/NADPH assay
NADP+ and NADPH levels were measured using NADP/NADPH-Glo assay (G9062, Promega). Cells were lysed by adding 200 μL/well of 0.1 mol/L NaOH with 0.5% DTAB and subjected to measurement in a Tecan Spark 10 M plate reader according to manufacturer’s instructions. The NADP+ and NADPH levels were normalized to cellular protein concentrations quantified by Bradford assay (Bio-Rad).
NADH/NADPH dehydrogenase activity assay
The NADH/NADPH dehydrogenase activity was determined as described previously36. The decrease in absorbance at OD340 nm was measured at room temperature in the substrate solution containing 250 μM NADH or NADPH in air-saturated 50 mM Tris-HCl, pH 8.0.
GR activity assay
The GR activity was measured using Glutathione Reductase Assay Kit (ab83461, Abcam). Samples were homogenized in cold assay buffer followed by adding 5 μL 3% H2O2 and incubated at 25 °C for 5 min to destroy GSH. Then 5 μL of catalase was added and incubated at 25 °C for another 5 min. The increase in absorbance at OD405 nm was measured at room temperature in Tecan Spark 10 M plate reader according to manufacturer’s instructions.
TrxR activity assay
TrxR activity was measured using Thioredoxin Reductase Activity Colorimetric Assay Kit (ab83463, Abcam). 2 × 106 cells were homogenized in 200 μL of cold TrxR assay buffer in the presence of the protease inhibitor on ice. After homogenization and centrifugation at 10,000 g for 15 minutes at 4 °C, the resulting supernatant was used for the assay. The total TrxR activity was measured in the presence of TrxR assay buffer (10 μL) via determining the total DTNB reduction, meanwhile, the background enzyme activity was measured in the presence of TrxR inhibitor (10 μL). The reaction kinetics were measured by recording the optical density (OD) at 412 nm using Tecan Spark 10 M plate reader at room temperature, according to manufacturer’s instructions.
Protein thermal shift assay
The Protein Thermal Shift Assay was conducted using a high-throughput approach on the Bio-Rad CFX Real-Time PCR Systems to measure protein stability57. Purified proteins were utilized at a final concentration of 0.5 mg/mL. During the assay, the proteins were incubated with SYPRO Orange Protein Gel Stain along with various buffer systems containing different concentrations of sodium chloride or magnesium chloride salts (0, 50, 100, and 200 mM). Additionally, substrates with NADH or NADPH were included at concentrations of 0, 50, 100, and 200 μM. The protein melt curves were obtained by subjecting the samples to a temperature range of 10 °C to 95 °C in 0.5 °C increments for 10 seconds. The Tm values were then determined based on these protein melt curves.
Nissl staining
Mice were perfused with PBS and 4% paraformaldehyde (Sigma). Brains were harvested, postfixed, and cryoprotected in 30% sucrose. Free-floating sections (30 μm) were cut via Leica C3050S Cryostat and mounted for Nissl staining as previously described36.
Molecular docking
The structures of the full-length AIF and AIF3 were predicted using ColabFold software (https://github.com/sokrypton/ColabFold). AIF(3) structures and NADPH were docked using CB-Dock2 (https://cadd.labshare.cn/cb-dock2/php/index.php) program. Docking models of mature AIF/NADPH and AIF3/NADPH were viewed and labeled via Pymol.
ICV injection assay
Two microliters of AAV-NDI1 or AAV-eGFP (1 × 1013 GC/mL) were injected into both sides of intracerebroventricular of newborn WT or AIF3 splicing mice as previously described56,58. The expression of NDI1 and eGFP was determined by immunostaining at postnatal day 21 or 60.
Quantification of protein dimerization via TwoMP analysis
Mass photometry experiments were conducted using a Refeyn TwoMP system and analyzed using the AcquireMP software. Microscope coverslips were pre-treated by rinsing both sides twice sequentially with Milli-Q water, isopropanol, and Milli-Q water. Excess water was removed using a nitrogen gun. For contrast-to-mass calibration, BSA was used as a standard. Immediately before measurements, protein stocks were diluted to 100 nM in buffer containing 25 mM HEPES (pH 7.4), 150 mM NaCl, and 1 mM EDTA. A volume of 1.8 µl of purified AIF or AIF3 protein solution was added to 16.2 µl of buffer and mixed gently by pipetting. Each sample was recorded for 60 seconds.
Statistical analysis
Statistical evaluation was performed by unpaired two-tailed Student’s t test between two groups and by one- or two-way ANOVA with Tukey’s, Dunnett’s, or Sidak’s multiple comparisons within multiple groups using GraphPad Prism 10.0 software. All representative images were repeated at least three times independently with similar results. The precise sample number (n) was provided to indicate the number of biological samples in each experiment. Immunoblots were quantified for statistical analysis, and the results were shown in the Source Data. Data are shown as mean ± SEM unless otherwise indicated. p < 0.05 is considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. The RNA-seq data were deposited at the Gene Expression Omnibus with accession number GSE261414. Source data for the figures and Supplementary Figs. are provided as a Source Data file. Source data are provided with this paper.
References
Ardissone, A. et al. A slowly progressive mitochondrial encephalomyopathy widens the spectrum of AIFM1 disorders. Neurology 84, 2193–2195 (2015).
Bano, D. & Prehn, J. H. M. Apoptosis-inducing factor (AIF) in physiology and disease: the tale of a repented natural born killer. EBioMedicine 30, 29–37 (2018).
Diodato, D. et al. A novel AIFM1 mutation expands the phenotype to an infantile motor neuron disease. Eur. J. Hum. Genet 24, 463–466 (2016).
Ghezzi, D. et al. Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor. Am. J. Hum. Genet 86, 639–649 (2010).
Hu, B. et al. A novel missense mutation in AIFM1 results in axonal polyneuropathy and misassembly of OXPHOS complexes. Eur. J. Neurol. 24, 1499–1506 (2017).
Kettwig, M. et al. From ventriculomegaly to severe muscular atrophy: expansion of the clinical spectrum related to mutations in AIFM1. Mitochondrion 21, 12–18 (2015).
Miyake, N. et al. X-linked hypomyelination with spondylometaphyseal dysplasia (H-SMD) associated with mutations in AIFM1. Neurogenetics 18, 185–194 (2017).
Morton, S. U. et al. AIFM1 mutation presenting with fatal encephalomyopathy and mitochondrial disease in an infant. Cold Spring Harb. Mol. Case Stud. 3, a001560 (2017).
Rinaldi, C. et al. Cowchock syndrome is associated with a mutation in apoptosis-inducing factor. Am. J. Hum. Genet 91, 1095–1102 (2012).
Sancho, P. et al. A newly distal hereditary motor neuropathy caused by a rare AIFM1 mutation. Neurogenetics 18, 245–250 (2017).
Sevrioukova, I. F. Structure/function relations in AIFM1 variants associated with neurodegenerative disorders. J. Mol. Biol. 428, 3650–3665 (2016).
Qiu, Y. et al. AIFM1 variants associated with auditory neuropathy spectrum disorder cause apoptosis due to impaired apoptosis-inducing factor dimerization. J. Zhejiang Univ. Sci. B 24, 172–184 (2023).
Klein, J. A. et al. The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature 419, 367–374 (2002).
Joza, N. et al. The molecular archaeology of a mitochondrial death effector: AIF in Drosophila. Cell Death Differ. 15, 1009–1018 (2008).
Cheung, E. C. et al. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. Embo J. 25, 4061–4073 (2006).
Pospisilik, J. A. et al. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell 131, 476–491 (2007).
Troulinaki, K. et al. WAH-1/AIF regulates mitochondrial oxidative phosphorylation in the nematode Caenorhabditis elegans. Cell Death Discov. 4, 2 (2018).
Wang, Y., Dawson, V. L. & Dawson, T. M. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp. Neurol. 218, 193–202 (2009).
Mate, M. J. et al. The crystal structure of the mouse apoptosis-inducing factor AIF. Nat. Struct. Biol. 9, 442–446 (2002).
Otera, H., Ohsakaya, S., Nagaura, Z., Ishihara, N. & Mihara, K. Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 24, 1375–1386 (2005).
Yu S. W., et al. Outer mitochondrial membrane localization of apoptosis-inducing factor: mechanistic implications for release. ASN Neuro. 1, e00021 (2009).
Miramar, M. D. et al. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J. Biol. Chem. 276, 16391–16398 (2001).
Churbanova, I. Y. & Sevrioukova, I. F. Redox-dependent changes in molecular properties of mitochondrial apoptosis-inducing factor. J. Biol. Chem. 283, 5622–5631 (2008).
Elguindy, M. M. & Nakamaru-Ogiso, E. Apoptosis-inducing Factor (AIF) and Its Family Member Protein, AMID, Are Rotenone-sensitive NADH:Ubiquinone Oxidoreductases (NDH-2). J. Biol. Chem. 290, 20815–20826 (2015).
Seo, B. B., Marella, M., Yagi, T. & Matsuno-Yagi, A. The single subunit NADH dehydrogenase reduces generation of reactive oxygen species from complex I. FEBS Lett. 580, 6105–6108 (2006).
Fang, J. & Beattie, D. S. External alternative NADH dehydrogenase of Saccharomyces cerevisiae: a potential source of superoxide. Free Radic. Biol. Med. 34, 478–488 (2003).
Park, J. S., Li, Y. F. & Bai, Y. Yeast NDI1 improves oxidative phosphorylation capacity and increases protection against oxidative stress and cell death in cells carrying a Leber’s hereditary optic neuropathy mutation. Biochim Biophys. Acta. 1772, 533–542 (2007).
Carneiro, P., Duarte, M. & Videira, A. Disruption of alternative NAD(P)H dehydrogenases leads to decreased mitochondrial ROS in Neurospora crassa. Free Radic. Biol. Med. 52, 402–409 (2012).
Vahsen, N. et al. AIF deficiency compromises oxidative phosphorylation. EMBO J. 23, 4679–4689 (2004).
Davidson, J. F. & Schiestl, R. H. Mitochondrial respiratory electron carriers are involved in oxidative stress during heat stress in Saccharomyces cerevisiae. Mol. Cell Biol. 21, 8483–8489 (2001).
Jimenez-Gomez, B. et al. Transgenic NADH dehydrogenase restores oxygen regulation of breathing in mitochondrial complex I-deficient mice. Nat. Commun. 14, 1172 (2023).
McElroy, G. S. et al. NAD+ regeneration rescues lifespan, but not ataxia, in a mouse model of brain mitochondrial complex i dysfunction. Cell Metab. 32, 301–308.e306 (2020).
Hangen, E. et al. A brain-specific isoform of mitochondrial apoptosis-inducing factor: AIF2. Cell Death Differ. 17, 1155–1166 (2010).
Delettre, C. et al. AIFsh, a novel apoptosis-inducing factor (AIF) pro-apoptotic isoform with potential pathological relevance in human cancer. J. Biol. Chem. 281, 6413–6427 (2006).
Delettre, C. et al. Identification and characterization of AIFsh2, a mitochondrial apoptosis-inducing factor (AIF) isoform with NADH oxidase activity. J. Biol. Chem. 281, 18507–18518 (2006).
Liu, S. et al. AIF3 splicing switch triggers neurodegeneration. Mol. Neurodegener. 16, 25 (2021).
Brosey, C. A. et al. Defining NADH-driven allostery regulating apoptosis-inducing factor. Structure 24, 2067–2079 (2016).
Ighodaro, O. M. & Akinloye, O. A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 54, 287–293 (2018).
Lee, W. L., Huang, J. Y. & Shyur, L. F. Phytoagents for cancer management: regulation of nucleic acid oxidation, ROS, and related mechanisms. Oxid. Med Cell Longev. 2013, 925804 (2013).
Berkholz, D. S., Faber, H. R., Savvides, S. N. & Karplus, P. A. Catalytic cycle of human glutathione reductase near 1 A resolution. J. Mol. Biol. 382, 371–384 (2008).
Iwata, M. et al. The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates. Proc. Natl Acad. Sci. USA 109, 15247–15252 (2012).
He, Y. et al. Mitochondrial complex I defect induces ROS release and degeneration in trabecular meshwork cells of POAG patients: protection by antioxidants. Invest Ophthalmol. Vis. Sci. 49, 1447–1458 (2008).
Monostori, P., Wittmann, G., Karg, E. & Turi, S. Determination of glutathione and glutathione disulfide in biological samples: an in-depth review. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 877, 3331–3346 (2009).
Yang, M. et al. KDM6B promotes PARthanatos via suppression of O6-methylguanine DNA methyltransferase repair and sustained checkpoint response. Nucleic Acids Res. 50, 6313–6331 (2022).
Wang Y., et al. KDM6B cooperates with Tau and regulates synaptic plasticity and cognition via inducing VGLUT1/2. Mol Psychiatry 12, 5213–5226 (2022).
Challis, R. C. et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc. 14, 379–414 (2019).
Luo, M. et al. ZMYND8 is a master regulator of 27-hydroxycholesterol that promotes tumorigenicity of breast cancer stem cells. Sci. Adv. 8, eabn5295 (2022).
Zhang, B. et al. Targeting BCAT1 Combined with alpha-Ketoglutarate Triggers Metabolic Synthetic Lethality in Glioblastoma. Cancer Res. 82, 2388–2402 (2022).
Zhang, L. et al. Mitochondrial localization of the Parkinson’s disease related protein DJ-1: implications for pathogenesis. Hum. Mol. Genet 14, 2063–2073 (2005).
Vaseva, A. V. & Moll, U. M. Identification of p53 in mitochondria. Methods Mol. Biol. 962, 75–84 (2013).
Rustin, P., Munnich, A. & Rotig, A. Succinate dehydrogenase and human diseases: new insights into a well-known enzyme. Eur. J. Hum. Genet 10, 289–291 (2002).
Bourgeron, T. et al. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet 11, 144–149 (1995).
Thorburn, D. R., Chow, C. W. & Kirby, D. M. Respiratory chain enzyme analysis in muscle and liver. Mitochondrion 4, 363–375 (2004).
Kirby, D. M., Thorburn, D. R., Turnbull, D. M. & Taylor, R. W. Biochemical assays of respiratory chain complex activity. Methods Cell Biol. 80, 93–119 (2007).
Fernandez-Vizarra, E. & Zeviani, M. Blue-native electrophoresis to study the OXPHOS complexes. Methods Mol. Biol. 2192, 287–311 (2021).
Wang Y., et al. A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science 354, aad6872 (2016).
Layton, C. J. & Hellinga, H. W. Quantitation of protein-protein interactions by thermal stability shift analysis. Protein Sci. 20, 1439–1450 (2011).
Ruan, Z. et al. MIF promotes neurodegeneration and cell death via its nuclease activity following traumatic brain injury. Cell Mol. Life Sci. 79, 39 (2021).
Acknowledgements
We thank Children’s Health and Pathology department at UT Southwestern Medical Center for providing de-identified human autopsy brain tissues with or without mitochondrial disorders, the UTSW Electron Microscopy Core for TEM sample preparation and the assistance with electron microscope images (1S10OD021685-01A1); Macromolecular Biophysics Resource Core for TwoMP analysis (S10OD030312-01); the Next Generation Sequencing Core for sequencing, and Metabolomics Core for metabolic analysis. This work was supported by grants from National Institutes of Health (R01AG066166, R01AG079094 and R00NS078049) and Darrell K Royal Research Fund to Y.W.
Author information
Authors and Affiliations
Contributions
M.Zhou: Formal analysis, investigation, visualization, methodology, writing original draft and editing; S.Q.L.: maintaining mouse lines, in vivo mouse brain investigation, neuron culture, ROS imaging; Y.N. Wang: cloning, virus preparation; B. Zhang: metabolism; M.Zhu: cloning and cell culture; J.E.W.: maintaining mouse lines; V.R. and Y.S. Fang: providing deidentified human brain tissues; W.L.: Formal analysis, supervision, visualization, writing original draft and editing; and Y.W.: Conceptualization, formal analysis, supervision, funding acquisition, visualization, writing original draft and editing, project administration.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Liu Yang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhou, M., Liu, S., Wang, Y. et al. AIF3 splicing variant elicits mitochondrial malfunction via the concurrent dysregulation of electron transport chain and glutathione-redox homeostasis. Nat Commun 16, 1804 (2025). https://doi.org/10.1038/s41467-025-57081-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-57081-5