Abstract
Skeletal muscle undergoes many alterations with aging. However, the impact of aging on muscle’s ability to secrete myokines and its subsequent effects on the body remain largely unexplored. Here, we identify myokines that have the potential to ameliorate age-related muscle and bone decline. Notably, circulating levels of cardiotrophin-like cytokine factor 1 (CLCF1) decrease with age, while exercise significantly upregulates CLCF1 levels in both humans and rodents. Restoring CLCF1 levels in aged male mice improves their physical performance, glucose tolerance, and mitochondrial activity. Furthermore, CLCF1 protects against age-induced bone loss by inhibiting osteoclastogenesis and promoting osteoblast differentiation in aged male mice. These improvements mirror some of the effects of exercise training. Conversely, blocking CLCF1 activity significantly abolishes these beneficial effects, confirming the crucial role of CLCF1 in mediating the positive effects of exercise on muscle and bone health in male mice. These findings collectively suggest that CLCF1 may contribute to the regulation of age-associated musculoskeletal deterioration, and warrant further investigation into its potential role as a modulator of musculoskeletal health during aging.
Similar content being viewed by others
Introduction
Aging leads to significant alterations in the musculoskeletal system and affects the bones, muscles, joints, and connective tissues, resulting in reduced function and mobility. However, regular physical activity has been shown to counteract these age-related changes, promoting better musculoskeletal health and overall quality of life. Muscles and bones share a mutual dependence, and their health is interconnected1. Several studies have demonstrated a correlation between sarcopenia and increased susceptibility to osteoporosis2. Notably, myokines have been shown to orchestrate intricate communication between muscle and bone, and play a key role in combating age-related decline in muscle and bone3,4. For example, exercise-induced myokine irisin exhibits a decrease in plasma level in aged mice5,6,7. Irisin supplementation induced muscle hypertrophy and enhanced skeletal muscle regeneration through satellite cell activation and expansion8, and promoted bone formation9,10. Furthermore, exercise reversed the aging-induced decrease in the level of the myokine apelin11. Apelin reversed age-related decline in skeletal muscle by enhancing mitochondrial biogenesis, counteracting inflammation, and stimulating muscle stem cells, and promoted osteogenic differentiation via the Wnt/β-catenin signaling pathway12. However, the question of how aging affects muscle’s ability to secrete myokines and the implications of these changes remain largely unexplored.
Considering that exercise-induced myokines are crucial for maintaining muscle function, we speculated that aged skeletal muscles may have a diminished capacity to produce these essential signaling molecules, potentially compromising muscle strength and regeneration. To identify exercise-induced myokines in trained old adult muscle tissue, we analyzed human skeletal muscle transcriptome datasets generated from exercise response studies in young and old adults13. Here, we identified cardiotrophin-like cytokine factor 1 (CLCF1) as an exercise-induced myokine that declines with aging. CLCF1, a member of the interleukin-6 (IL-6) family, has been identified as a regulator of the glycoprotein 130 (gp130)/leukemia inhibitory factor receptor (LIFRβ) signaling pathway. It functions by binding to the ciliary neurotrophic factor receptor (CNTFR), promoting the recruitment and phosphorylation of gp130 and LIFR14. Additionally, CLCF1 may engage alternative receptor mechanisms that remain unidentified15,16. CLCF1 plays several roles in development, motor neuron survival, immunomodulatory functions, and cancer17,18,19,20. Mutations in CLCF1 are associated with crisponi/cold-induced sweating syndrome (CS/CISS) with dysregulated body temperature, facial muscle weakness, and skeletal abnormalities21,22. However, the role of CLCF1 in skeletal muscle and bone remains unclear. In this study, we investigated the role of CLCF1 in mediating exercise-induced benefits. We observed that elevating CLCF1 levels significantly enhanced muscle function and bone formation, while blocking CLCF1 abolished these exercise-induced effects.
Results
Exercise induces CLCF1 production in skeletal muscle
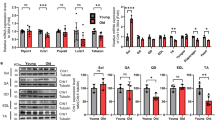
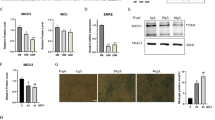
We hypothesized that the aging-related decline in muscle function could be attributed to reduced secretion of myokines from aging muscle. To identify myokines affected by aging, we aimed to analyze the changes in secreted protein profiles in muscle tissues from young and old individuals before and after exercise. We reanalyzed transcriptomic data from muscle biopsies obtained in a previous study by Raue et al.13. This study investigated the effects of acute resistance exercise (RE), progressive resistance training (PRT), and aging on gene expression in human skeletal muscle. Two groups were examined: young adults (24–25 years old) and older adults (78–84 years old). Both age groups participated in a 12-week PRT program, with muscle biopsies collected from the vastus lateralis muscle before and after a single RE session, as well as before the first and last sessions of the PRT program. Biopsies were also collected from untrained young and older adults to analyze baseline differences. We utilized the dataset from mixed muscle containing both slow and fast-twitch fibers (GSE28422). We screened 2933 genes encoding secreted proteins, focusing on those that are altered in trained muscles (Supplementary Data 1 and 2). Specifically, we aimed to identify exercise-inducible genes that do not exhibit a response to a single bout of exercise in older individuals but show alterations with chronic exercise. Our analysis revealed significant alterations in the mRNA expression of cardiotrophin-like cytokine factor 1 (CLCF1), lecithin-cholesterol acyltransferase (LCAT), C-C motif chemokine ligand 2 (CCL2), and probable threonine protease PRSS50 (PRSS50) in older adults following a 12-week exercise program (Fig. 1A, B and Fig. S1A, B). Among the significantly upregulated secretory genes, CLCF1 emerged as a promising candidate due to its exercise-induced upregulation in muscle tissue23,24,25,26. Despite the lack of known functional roles for CLCF1, the expression of its receptors (gp130/LIFR/CNTFR) is altered in skeletal muscles undergoing atrophy, suggesting a potential link between CLCF1 and muscle health27,28,29. Following the identification of CLCF1 as a muscle-derived exerkine, we systematically investigated its plasma concentration in healthy young and older adults before and after exercise under various experimental conditions. In young adults (human study 1) and young jiu-jitsu athletes (human study 2), both resistance and high-intensity interval exercise performed for one hour led to a significant increase in circulating plasma CLCF1 levels (Fig. S1C, D). The rigorous training regimen, characterized by high intensity and frequency, coupled with advanced training experience and competitive participation, may account for the elevated baseline CLCF1 levels in the young jiu-jitsu athletes group. However, treadmill aerobic exercise for one hour did not significantly increase plasma CLCF1 levels (human study 3) (Fig. S1E). We further demonstrate that both resistance training for 16 weeks (human study 4) and resistance band exercise for 12 weeks (human study 5) significantly elevated plasma CLCF1 levels in older adults (Fig. S1F, G). Additionally, we investigated differences in plasma CLCF1 levels between young and elderly groups under non-exercise conditions. For this analysis, we analyzed levels of CLCF1 in plasma samples from subjects in Fig. S1C–G and additionally recruited participants (human study 6) under non-exercise conditions. Our data revealed that circulating CLCF1 levels were significantly lower in elderly individuals compared to young subjects (Fig. S1H). These findings are exploratory and require further validation in larger and more controlled studies. Based on these results, we investigated whether CLCF1 is mainly produced by skeletal muscle. C2C12 myotubes were treated with AICAR, an AMPK activator, which is one of the signaling pathways activated during exercise, and the protein and mRNA levels of CLCF1 were examined. AICAR treatment significantly increased both CLCF1 protein and mRNA expression (Fig. 1C, D). We further confirmed that electrical pulse stimulation (EPS), an in vitro exercise model that mimics muscle adaptations in vivo, increased CLCF1 expression (Fig. 1E, F). Furthermore, high-intensity treadmill running significantly upregulated CLCF1 mRNA expression specifically within the muscle tissue of young mice. Importantly, no significant differences in CLCF1 mRNA expression were observed in other tissues examined both before and after exercise (Fig. 1G). Consistent with these findings, plasma CLCF1 concentrations significantly increased after a single bout of exercise in young mice, whereas exercise-induced CLCF1 release was not significantly elevated in older mice (Fig. 1H). Interestingly, under basal conditions, plasma CLCF1 levels were significantly lower in old mice compared to young mice. To investigate this further, we measured CLCF1 levels in mice at 3, 8, and 18 months of age, and observed a progressive decline in CLCF1 levels with increasing age (Fig. 1I). Moreover, we investigated CLCF1 expression in muscle tissue throughout the aging process using SarcoAtlas (https://sarcoatlas.scicore.unibas.ch), a comprehensive resource for muscle gene expression data. Our analysis demonstrated that CLCF1 expression remained stable with age, whereas cytokine receptor-like factor 1 (CRLF1), a protein crucial for CLCF1 secretion, exhibited a significant decline in expression (Fig. S2A, B). In line with these findings, we observed a substantial reduction in Crfl1 expression in aged mouse muscles compared to young muscles. Conversely, Clcf1 expression showed no significant age-related alterations (Fig. S2C). These results suggest that the age-associated decrease in CLCF1 plasma levels may be primarily attributed to the downregulation of CRLF1. To elucidate whether the low plasma CLCF1 levels in exercised aged mice were attributed to reduced CRLF1 expression, we analyzed the expression changes of CLCF1 and CRLF1 in the skeletal muscles of aged mice following exercise. Unlike young mice, aged mice demonstrated no significant upregulation of CLCF1 and CRLF1 expression following exercise. The observed absence of CLCF1 and CRLF1 upregulation in aged mice following exercise might be attributable to age-related alterations in cellular signaling, receptor sensitivity, or transcriptional regulation (Fig. S2D). Collectively, these results indicate that aging may affect the production of the exercise-induced CLCF1.
A Volcano plots showing differentially expressed genes (DEGs) encoding secretory proteins in skeletal muscles of young and old adults before and after 12 weeks of progressive resistance training. B Expression levels of genes commonly found in three DEG groups (yBase, n = 15; y1st, n = 16; yFinal, n = 16; oBase, n = 12; o1st, n = 12; oFinal, n = 12, p = 0.0278, 0.0453, 0.0223 (CLCF1); 0.0011, 0.0379 (LCAT); 0.005, 0.0299, 0.014, 0.0095 (CCL2); 2.00e-7, 0.0002 (PRSS50): young and old adults). C Clcf1 expression in C2C12 myotubes treated with AICAR (1 mM, 24 h; n = 3, p = 0.001; biological replicates). D Left: Immunoblot analysis of C2C12 myotubes treated with AICAR. Right: Quantification by densitometry analysis (n = 4, p = 0.0026). E Clcf1 expression in C2C12 myotubes subjected to electrical pulse contraction (EPS; n = 6, p = 0.0015; biological replicates). F Left: Immunoblot of C2C12 myotubes after EPS. Right: Quantification of western blots by densitometry analysis (n = 3, p = 0.046). G mRNA expression of Clcf1 in skeletal muscles following treadmill running (3 mo, male, created in BioRender.com) (n = 4; p = 0.0062, 0.047, 0.018). H Plasma concentration of CLCF1 measured 30, 60, or 90 min after treadmill exercise in young (3 mo, male) and aged (24 mo, male) mice (n = 4, p = 0.026, 0.038, 1.3e-4), Box plots show the median (center line), interquartile range (25th–75th percentiles), and the full data range (minimum to maximum, shown as whiskers). I Plasma CLCF1 concentration in young (3 mo, male), middle-aged (8 mo, male), and aged (18 mo, male) mice (n = 10; p = 3.4.e-4, 1e-6, 0.041). Data are presented as means ± s.e.m.; one-way ANOVA with post hoc Tukey’s multiple comparison test (B, I); two-tailed unpaired Student’s t-test (C–G); two-way ANOVA with post hoc Tukey’s multiple comparison test (H). *p < 0.05, **p < 0.005 and ***p < 0.001. Source data are provided as a Source Data file.
CLCF1 supplementation ameliorates age-related muscle weakness and bone loss
CLCF1 receptors (gp130/LIFRβ/CNTFRa) are upregulated in muscle tissues in models of muscle atrophy27,28,29. We hypothesized that exercise might modulate the expression of these receptors as a potential adaptive response to muscular stress. Consistent with this hypothesis, physical exercise resulted in increased expression of Gp130, Lifr, and Cntfr in the skeletal muscle of mice (Fig. S3A). Given the observed changes in young mice, we further investigated whether aging might impair this adaptive response mechanism. Conversely, aged mice demonstrated a notable decrease in Gp130 and Lifr expression within muscle tissues (Fig. S3B). These observations led us to hypothesize that CLCF1 plays a critical role in muscle function and is essential for maintaining skeletal muscle homeostasis. To investigate the effects of CLCF1 on muscle health, aged male mice received intraperitoneal injections of recombinant CLCF1 at two different doses: 0.1 and 0.5 mg/kg (Fig. 2A). Plasma CLCF1 concentrations increased after CLCF1 injection at 0.5 mg/kg doses (Fig. S4A). There were no statistically significant differences in muscle, heart, and body weights between the control and CLCF1 treatment groups (Fig. 2B, Fig. S4B, C). Mice administered 0.1 mg/kg of CLCF1 exhibited no significant alterations in grip strength compared to control aged mice. In contrast, when the CLCF1 dosage was increased to 0.5 mg/kg, a statistically significant enhancement in grip strength was observed (Fig. 2C). In the running test, aged mice injected with both 0.1 and 0.5 mg/kg of CLCF1 demonstrated a statistically significant improvement in running endurance compared to control aged mice. Notably, mice administered 0.5 mg/kg exhibited a higher proportion of individuals reaching maximal running time compared to the 0.1 mg/kg group (Fig. 2D–F). Histological analysis showed that CLCF1 treatment significantly increased the cross-sectional area of TA muscle fibers without affecting muscle fiber number, compared to the control group. (Fig. 2G–K). Although there was no significant difference in twitch force potentiation between the groups (Fig. 2L), tetanic force was significantly higher in CLCF1-treated muscles than in the muscles of aged mice in the control group (Fig. 2M). Additionally, CLCF1 treatment enhanced fatigue resistance in aged mice (Fig. 2N, O). Next, we analyzed the molecular changes in the muscle of aged mice following CLCF1 administration (0.5 mg/kg). Real-time polymerase chain reaction (RT-qPCR) was performed to examine the expression of MyHC isoforms, markers of muscle fiber type, in the mouse skeletal muscle. In the soleus muscle of aged mice, the expression levels of Myh7 and Myh2 were significantly elevated; however, these increases were markedly attenuated by CLCF1 treatment. In contrast, the expression of Myh4 mRNA was significantly reduced in aged mice compared to young controls. CLCF1 treatment in aged mice restored Myh4 (type 2B) expression to levels comparable to those observed in young mice, although the increase did not reach statistical significance (Fig. S4D, E). CLCF1 treatment enhanced the phosphorylation of key proteins involved in muscle anabolic signaling pathways, including AKT, mTOR, S6K, and 4E-BP1 (Fig. S4F, G).
A Experimental scheme (created in BioRender.com). Aged mice (20 mo, male) were treated daily with CLCF1 (0.1 or 0.5 mg/kg) or vehicle for 2 weeks. B Muscle weights (SOL, GA, EDL, and TA) (vehicle, n = 8; 0.1 mg/kg, n = 9; 0.5 mg/kg, n = 11). C Grip strength test (vehicle, n = 11; 0.1 mg/kg, n = 7; 0.5 mg/kg, n = 10; p = 0.0081). D Running curves (vehicle, n = 8; 0.1 mg/kg, n = 7; 0.5 mg/kg, n = 7; p = 6.7e-5, 0.0018), E distance ran (vehicle, n = 8; 0.1 mg/kg, n = 8; 0.5 mg/kg, n = 7; p = 0.0008, 0.0012), and F time to exhaustion (vehicle, n = 8; 0.1 mg/kg, n = 8; 0.5 mg/kg, n = 7; p = 0.0010, 0.0014). G Left: Full cross-section of TA muscle. Scale bar, 0.5 mm. Right: Total muscle fiber number (vehicle, n = 6; 0.5 mg/kg, n = 5). H Laminin-stained myofibers in TA of young, old, and old-CLCF1 mice (0.5 mg/kg). Scale bar, 50 μm. I–K Quantification of muscle fiber size (n = 3, biological replicates): I mean muscle fiber area (p = 0.038), J frequency distribution of fiber areas, and K quartile analysis of the distribution (p = 0.0364, 0.0427, 0.0406). L The maximum twitch force at superamaximal voltage, M tetanic force at different stimulation frequencies (10–150 Hz) and area under curve (p = 0.039, 0.0043, 0.0040), N, O fatigue resistance and area under curve of old and old-CLCF1 mice (n = 10; p = 0.034). P Representative micro-CT images of the trabecular and cortical bone compartments of distal femoral metaphyseal regions (old and old-CLCF1 mice, 0.5 mg/kg) Scale bar, 1 mm. Q Measurement of trabecular bone volume/total volume (BV/TV), trabecular thickness (TbTh), trabecular separation (TbSp), trabecular number (TbN), cortical bone volume/total volume (Corti BV/TV), and cortical thickness (Corti Th) (n = 8; p = 0.022, 0.196, 0.038, 0.013, 0.019, 0.093). R Representative hematoxylin/eosin (H&E), TRAP, and Runx2-stained histological sections of proximal tibiae. Scale bar, 100 μm. S Quantification of osteoclast number per bone surface (n = 8; p = 0.02) and T Osteoblast (n = 8). Data are presented as means ± s.e.m.; two-tailed unpaired Student’s t-test (B–K, O, Q, S, T); Mann–Whitney U test (D); or two-way ANOVA with post hoc Bonferroni’s multiple comparison test (M, N). *p < 0.05, **p < 0.005 and ***p < 0.001; n.s., p > 0.05. Source data are provided as a Source Data file.
Currently, there are discrepancies on the role of CLCF1 in bone health16,30, indicating the need for further research on the association between CLCF1 expression and bone health. Exercise is widely recognized as one of the most effective interventions for improving and maintaining bone health, primarily through its significant impact on the muscle-bone unit. Given the critical role of muscle-derived factors in bone metabolism and the existing knowledge gaps in this ___domain, we hypothesized that CLCF1 might exert beneficial effects on bone health. To address this hypothesis, we subsequently investigated the potential protective role of CLCF1 in maintaining bone integrity and elucidated its underlying molecular mechanisms. We found that CLCF1 treatment (0.5 mg/kg) significantly increased bone volume and decreased trabecular separation in the trabecular bone of aged mice. Additionally, CLCF1 treatment significantly increased both the volume and thickness of the cortical bone (Fig. 2P, Q). Histological analysis revealed that CLCF1 treatment influenced bone remodeling in aged mice by regulating changes in osteoclast and osteoblast activities. Specifically, CLCF1 treatment significantly reduced the number of osteoclasts and slightly increased the number of osteoblasts, although the effect was not significant. (Fig. 2R–T).
We next investigated whether CLCF1’s beneficial effects extend to aged female mice. Our study demonstrated that CLCF1 significantly improved muscle function (Fig. S5A–H) and inhibited bone deterioration (Fig. S5I, J) in these mice. Collectively, these data demonstrate that CLCF1 exerts potent inhibitory effects on osteoclast activity while modestly stimulating osteoblast function, thereby promoting increased bone mass.
To evaluate whether CLCF1 exerts similar effects in young mice, we administered a 0.5 mg/kg dose of CLCF1 to 3-month-old mice for two weeks and subsequently analyzed their muscle and bone parameters (Fig. S6A). A significant increase in soleus muscle weight was observed, while no differences were detected in other muscles and body weight (Fig. S6B, C). However, no significant differences were observed in grip strength or running performance between the groups (Fig. S6D–F). Notably, a higher proportion of CLCF1-injected mice ran to the maximum time. No statistically significant differences were observed in bone parameters between CLCF1-injected and control mice (Fig. S6G, H). These findings suggest that CLCF1 levels are already within the normal physiological range in young mice. Given the optimal muscle and bone function typically observed at this age, the effects of CLCF1 treatment are less pronounced compared to older mice, where there is a greater capacity for improvement in muscle and bone health.
CLCF1 treatment stimulates glucose uptake and improves glucose metabolism
Given that multiple signaling pathways, including AKT/mTOR, JAK/STAT, and ERK1/2, are known to be triggered by CLCF1 and are involved in muscle function31,32, we hypothesized that CLCF1 may modulate key cellular signaling mechanisms in muscle cells. To test this hypothesis, we investigated the effects of CLCF1 on the expression and activation of these signaling pathways in C2C12 cells. CLCF1 treatment activated AKT/mTOR/S6K, STAT3, and ERK1/2 pathways, but inactivated the forkhead box protein O 1/3 (FoxO1/3) signaling pathway (Fig. S7A, B). Notably, previous literature suggests that STAT3 activation can promote mitochondrial respiration, depending on the specific cellular context33,34,35. To explore whether CLCF1 enhances mitochondrial respiration, we measured oxygen consumption rate (OCR) in CLCF1- and vehicle-treated myotubes. CLCF1 treatment significantly increased basal, ATP-linked, and maximal respiration in myotubes compared with that in the control group (Fig. 3A, B). However, the effect of CLCF1 on mitochondrial respiration was markedly abrogated in the presence of the STAT3 inhibitor BP-1-102 (Fig. S8A). The tricarboxylic acid (TCA) cycle receives metabolites from various sources through glycolysis, fatty acid oxidation (FAO), and amino acid catabolism. To clarify the energy dependence of mitochondrial metabolism in CLCF1-treated cells, myotubes were treated with targeted inhibitors of glycolysis, FAO, and glutaminolysis. Glycolysis suppression had a significantly higher inhibitory effect on OCR in CLCF1-treated cells than FAO and glutaminolysis inhibition (Figs. 3C, D, S8B, C). Notably, CLCF1 treatment did not affect mitochondrial DNA copy number or the expression of genes involved in mitochondrial biogenesis (Fig. 3E, F). We next sought to determine whether the increased OCR observed following CLCF1 treatment was due to elevated glucose uptake and glycolytic activity. CLCF1 treatment significantly increased glucose uptake compared with that in the control group, both in the presence and absence of insulin, but did not affect fatty acid uptake (Fig. 3G, H). To investigate whether CLCF1 induces glucose uptake in different cell types, we analyzed glucose uptake and metabolic changes in 3T3-L1 adipocytes. CLCF1 effectively induced phosphorylation of STAT3 and AKT in 3T3-L1 adipocytes (Fig. S9A). CLCF1 stimulated glucose uptake in the absence of insulin but did not significantly enhance insulin-induced glucose uptake in 3T3-L1 adipocytes (Fig. S9B). These findings suggest that CLCF1 may modulate glucose metabolism not exclusively in muscle tissue, but potentially also in adipose tissue. In adipocytes, the limited effect of CLCF1 in the presence of insulin may be attributed to saturation of the insulin signaling pathway or full activation of metabolic enzymes and pathways by insulin. In addition, CLCF1 treatment did not affect the expression of genes involved in mitochondrial electron transport chain (ETC), glycolysis, and FAO. Additionally, no significant changes in OCR were observed upon CLCF1 treatment (Fig. S9C–D). To investigate to role of glucose transporters in CLCF1-induced increase in glucose uptake, we examined the expression of GLUT1 and GLUT4 in CLCF1-treated myotubes. Notably, CLCF1 treatment increased GLUT1 protein levels in the plasma membrane, but did not affect GLUT4 expression (Fig. 3I). Given the observed increase in glucose uptake following CLCF1 treatment, we hypothesized that this enhanced glucose influx would stimulate glycolysis. To test this, we measured extracellular acidification rate (ECAR) in C2C12 myotubes to directly assess the impact of CLCF1 on glycolytic metabolism. CLCF1 treatment significantly enhanced both glycolysis and glycolytic capacity in myotubes (Fig. 3J, K). Furthermore, the treatment upregulated several key glycolytic enzymes (Fig. 3L, M). Aged muscles exhibit a significant decline in both glycolytic enzyme expression and glycolytic capacity, which contributes to metabolic dysfunction during aging36,37. Given these age-related metabolic changes, we investigated whether CLCF1 might play a restorative role in reversing the age-associated decline of glycolytic enzyme expression. CLCF1 treatment specifically restored the decreased expression of PFKM and PKM2 in the muscle of aged mice (Fig. 3N). Based on these findings, to further evaluate the effects of CLCF1 treatment on glucose metabolism, we performed intraperitoneal glucose tolerance tests (GTT) and insulin tolerance tests (ITT) two weeks after CLCF1 injection. CLCF1 treatment significantly improved glucose tolerance but did not improve insulin tolerance in old mice (Fig. 3O–R). Overall, these results indicate that exercise-induced CLCF1 expression facilitates glycolysis and enhances glucose uptake by muscle cells, potentially contributing to glucose metabolism and muscle function in aged mice.
A, B Oxygen consumption rate (OCR) and quantification of basal respiration, ATP production, and maximal respiration in C2C12 myotubes treated with CLCF1 (100 ng/ml) for 48 h (vehicle, n = 10; CLCF1, n = 9; biological replicates, p = 1.0e-6, 7.0e-7, 1.3e-8). C OCR traces and D quantification in response to UK5099. (n = 10; p = 1.6e-5, 5.2e-7, 1.6e-4, 4.6e-5, 2.0e-11, 2.0e-11, 2.0e-11). E, F Mitochondrial DNA content and expression of mitochondrial genes in C2C12 myotubes after 48 h of CLCF1 treatment (n = 3). G Relative level of glucose uptake in C2C12 myotubes with CLCF1 treatment overnight (n = 6; p = 9.8e-3, 2.86e-2). H Relative level of fatty acid uptake in C2C12 myotube with CLCF1 overnight (n = 5). I Left: Western blot analysis of GLUT1 and GLUT4 in membrane and cytosolic fractions after 24 h of treatment with vehicle or CLCF1 in C2C12 myotubes (n = 3; p = 0.0469). Right: Quantification of Western blots was performed by densitometry analysis. J Measurement of glycolysis by extracellular acidification rate (ECAR) in C2C12 myotubes treated with vehicle or CLCF1 for 48 h (n = 8). K Quantification of glycolysis and glycolytic capacity (p = 3.0e-4, 2.0e-4). L Expression of glycolysis-related enzymes in C2C12 myotubes treated with vehicle or CLCF1 for 24 h. M Quantification of glycolytic enzymes (n = 4; p = 3.44e-2, 4.6e-3, 2.05e-2, 2.2e-3). N Top: Expression of glycolysis-related enzymes in muscle of young (3 mo, male), old (20 mo, male), and old-CLCF1 (20 mo, male). Bottom: Quantification of western blots (n = 4; p = 0.0313, 0.0135, 0.0049, 0.0001). O Intraperitoneal glucose tolerance test (IPGTT) in young, old, and old-CLCF1 (p = 4.8e-5, 7.5e-5). P Area under curve (AUC) (young, n = 12; old, n = 10; old_CLCF1, n = 8; biological replicates, p = 0.0149, 0.0293). Q Insulin tolerance test in young, old, and old-CLCF1 (p = 2.5e-6, 8.2e-7). R AUC young, n = 8; old, n = 11; old_CLCF1, n = 10; p = 0.0251, 0.0135. Data are presented as means ± s.e.m.; two-tailed unpaired Student’s t-test (B, E–I, K, M); one-way ANOVA with post hoc Bonferroni’s multiple comparison test (J, N, P, R); or two-way ANOVA with post hoc Tukey’s multiple comparison test (D, O, Q). *p < 0.05, **p < 0.005 and ***p < 0.001; n.s., p > 0.05. Source data are provided as a Source Data file.
CLCF1 inhibits bone resorption and enhances bone formation
Considering that CLCF1 contributes to increased bone mass through strong inhibition of osteoclast formation and slight increase in osteoblast formation in vivo, we investigated its direct effects on osteoclast and osteoblast differentiation in vitro. To assess the effect of CLCF1 on osteoclastogenesis, bone marrow-derived macrophages (BMMs) were treated with varying concentrations of CLCF1 during RANKL-induced osteoclast differentiation. CLCF1 treatment inhibited RANKL-induced osteoclast formation in a dose-dependent manner (Fig. 4A) and significantly suppressed the expression of key osteoclast-related genes at both the mRNA and protein levels (Fig. 4B, C). CLCF1 receptor components (Cntfr, Lifr, and Gp130) and their binding partner, cytokine receptor-like factor 1 (CRLF1), were expressed in bone and joint tissues, suggesting their potential involvement in skeletal and joint-related physiological and pathological38,39. These findings prompted us to investigate whether CLCF1 exerts a cell-autonomous regulatory effect on osteoclast differentiation. Although CLCF1 contains a signal sequence, its secretion is inefficient unless co-expressed with its chaperone, CRLF1. We confirmed that the CLCF1/CRLF1 complex produced in Expi293 cells induced signaling activation and increased oxygen consumption rate (OCR) in C2C12 cells, demonstrating a response similar to native CLCF1 treatment (Fig. S10A–D). Based on these preliminary findings, we co-overexpressed CLCF1 and CRLF1 in bone marrow macrophages (BMMs). Notably, this overexpression significantly inhibited osteoclast differentiation (Fig. 4D). To confirm the protective effects of CLCF1 against bone resorption in vivo, we established a model of RANKL-induced bone loss (Fig. 4E). Micro-CT analyses demonstrated the potent inhibitory effect of CLCF1 on RANKL-induced bone resorption in vivo (Fig. 4F–J). To confirm the effect of CLCF1 on osteoblastogenesis, osteoblast precursor cells were treated with CLCF1 during osteoblast differentiation. CLCF1 treatment slightly enhanced ALP activity, significantly stimulated nodule formation (Fig. 4K, L), and significantly upregulated the expression of the osteoblast marker genes Alpl and Ibsp (Fig. 4M). Next, we investigated whether CLCF1 autonomously regulates osteoblast differentiation and found that CLCF1 and CRLF1 overexpression in osteoblast precursor cells significantly enhanced osteoblast differentiation (Fig. 4N, O). Furthermore, we implanted BMP2-soaked sponges with or without CLCF1 into a dorsal subcutaneous pocket to evaluate CLCF1’s potential to stimulate bone formation in vivo. Implantation of sponges containing BMP2 with CLCF1 significantly enhanced BMP2-induced ectopic bone formation compared to the BMP2 with PBS control groups (Fig. 4P–R). Collectively, these results suggest that CLCF1 negatively regulates osteoclast differentiation and positively regulates osteoblast differentiation to increase bone mass.
A–C BMMs were treated with different concentrations of CLCF1 in the presence of M-CSF and RANKL for 3 days. A The numbers of TRAP-positive osteoclasts were counted (n = 4; biological replicates, p = 3.32e-5, 4.6e-9, 2.0e-10). Scale bar, 100 μm. B Real-time PCR analyses for the expression of the indicated genes (n = 3; p = 2.2e-7, 8.0e-4, 0.4408, 1.0e-9, 1.0e-9). C Western blot for the expression of the indicated molecules. D TRAP staining and quantification of osteoclasts differentiated from BMMs transduced with pMX-IRES-EGFP or CLCF1 and CRLF1 retrovirus for 3 days (n = 3; biological replicates, p = 0.0081). Scale bar, 100 μm. E Schemes of RANKL-induced bone loss (created in BioRender.com). F Representative µCT images of the trabecular and cortical compartments of distal femoral metaphyseal regions. Scale bar, 1 mm. G–J Measurement of trabecular bone volume /total volume (BV/TV, p = 5.0e-5, 0.0052, 0.019), trabecular thickness (TbTh, p = 0.0174, 0.0032), trabecular separation (TbSp, p = 0.0329, 0.0007), trabecular number (TbN, p = 0.0014, 0.000046) (n = 4). K–M Osteoblast precursors differentiated with OGM for 3 or 6 days in the presence or absence of CLCF1 (100 ng/mL). K ALP assay after 3 days (n = 3; biological replicates, p = 0.0130). L Alizarin red staining and quantification at 6 days (n = 3; p = 0.0208). M PCR analyses for the expression of the indicated genes (n = 3; p = 1.0e-4, 1.0e-10). N, O Osteoblasts transduced with pMX-IRES-EGFP or CLCF1 and CRLF1 retrovirus were differentiated with OGM for 3 or 6 days. N ALP activity was measured (n = 3; p = 0.0198). O Alizarin red staining and quantification (n = 3; p = 0.0064). P Schemes of BMP2-induced ectopic bone formation (created in BioRender.com). Q Representative µCT images of the ectopic bones. Scale bar, 1 mm. R Measurement of BV and BV/TV (n = 6; p = 0.0451, 0.0437). Data are presented as means ± s.e.m.; two-tailed unpaired Student’s t-test (A, D, R); one-way ANOVA with post hoc Tukey’s multiple comparison test (G–J); or two-way ANOVA with post hoc Tukey’s multiple comparison test (B, K–O). *p < 0.05, **p < 0.005 and ***p < 0.001. Source data are provided as a Source Data file.
To elucidate the underlying mechanisms through which CLCF1 regulates osteoclast and osteoblast differentiation, we examined the modulation of STAT signaling. Considering that CLCF1 can induce STAT1 and STAT3 phosphorylation, we investigated whether these pathways mediate the effects of CLCF1 in bone cells. CLCF1 pretreatment for 1 h markedly induced STAT1 and STAT3 phosphorylation in both BMMs and osteoblasts (Fig. S11A, D). Further experiments using STAT1- and STAT3-specific siRNAs revealed that STAT1 plays a key role in osteoclastogenesis regulation, whereas STAT3 is more critical for osteoblastogenesis. STAT1 knockdown significantly mitigated the inhibitory effect of CLCF1 on osteoclast formation (Fig. S11B, C), whereas STAT3 knockdown effectively abrogated the stimulatory effect of CLCF1 on osteoblast differentiation (Fig. S11E, F). Overall, these results suggest that CLCF1 exerts its anti-osteoclastogenic and pro-osteogenic functions via STAT1 and STAT3 phosphorylation, respectively.
Enhanced muscle function and bone formation in CLCF1 transgenic mice
To investigate the long-term effects of elevated circulating CLCF1 levels, we generated transgenic (TG) mice overexpressing CLCF1. Considering that CLCF1 secretion relies on a heteromeric complex with CRLF140, we constructed and expressed CLCF1 fused to CRLF1 for enhanced secretion (Fig. 5A). Expectedly, Clcf1 and Crlf1 mRNA expression and circulating CLCF1 levels were significantly higher in the muscle tissue of TG mice than in wild-type (WT) mice (Fig. 5B–D). While the mean weights of EDL, TA, and GA muscles showed a non-significant increasing trend in TG mice, soleus muscle mass was significantly higher in TG mice compared to WT mice (Fig. 5E). Additionally, forelimb grip strength, grid hang time, and running capacity were higher in TG mice than in WT mice (Fig. 5F–H). Histological analysis showed that TG mice exhibited a larger cross-sectional area of TA muscle fibers compared to WT mice, while muscle fiber number remained unchanged (Fig. 5I, J). As expected, TG mice exhibited improved glucose tolerance than WT mice (Fig. 5K).
A Schematic representation of CLCF1-CRLF1 transgenic mice generation (created in BioRender.com). Male TG mice (10 weeks) were used. B, C Relative mRNA expression levels of Clcf1 (WT, n = 9; TG, n = 8; p = 0.0038) (B) and Crlf1 (WT, n = 9; TG, n = 7; p = 0.0051) (C) in TA muscle. D Plasma levels of CLCF1 protein from control and TG mice (WT, n = 4; TG, n = 5; p = 0.0240). E Weight of dissected SOL, TA, EDL, and GA muscles (WT, n = 9; TG, n = 7; biological replicates, p = 0.0096). F Grip strength test (WT, n = 9; TG, n = 7; p = 6.0e-6). G Grid hanging test (WT, n = 8; TG, n = 9; p = 0.0252). H Treadmill running performance test in mice: running curves, distance run, and time to exhaustion (WT, n = 8; TG, n = 7; p = 0.0008, 0.004, 0.003). I The number of fibers in the TA muscle (n = 4; biological replicates). J Immunostaining for myofiber boundaries using laminin in TA muscles of WT and TG mice (WT, n = 3; TG, n = 3; p = 0.0201, 0.001, 6.2e-7, 0.014, 0.005, 7.6e-7). Scale bar, 50 μm. K Glucose tolerance test in WT and TG mice (n = 9; p = 0.0494, 0.0001, 0.0318). L Representative µCT images of the trabecular and cortical compartments of distal femoral metaphyseal regions of control and TG mice. Scale bar, 1 mm. M Measurement of trabecular bone volume/total volume (BV/TV), trabecular thickness (TbTh), trabecular separation (TbSp), trabecular number (TbN), cortical bone volume/total volume (Corti BV/TV), and cortical thickness (Corti Th) (WT, n = 9; TG, n = 8; p = 0.0071, 0.0349, 0.0461, 0.0134). N TRAP, hematoxylin/eosin (H&E), and Runx2 staining of a histological section of proximal tibiae. Scale bar, 100 μm. O Osteoclast number per bone surface and osteoblast number per bone surface were assessed (WT, n = 9; TG, n = 8; p = 0.0001, 0.0067). Data are presented as means ± s.e.m.; two-tailed unpaired Student’s t-test (B–O); Log-rank (Mantel–Cox) test (H); or two-way ANOVA with post hoc Bonferroni’s multiple comparison test (K). *p < 0.05, **p < 0.005 and ***p < 0.001. Source data are provided as a Source Data file.
To investigate the effects of CLCF1 overexpression on bone, micro-CT scans were performed on TG and WT mice. Compared with that in WT mice, there was a significant increase in trabecular bone volume, thickness, and number, and cortical bone volume and thickness in the TG mice. Additionally, trabecular separation was significantly reduced in the TG mice (Fig. 5L, M). Moreover, there was a significant decrease in osteoclast number and a significant increase in osteoblast number in the TG mice compared with that in WT mice (Fig. 5N, O). These results indicate that CLCF1 upregulation increases bone mass in vivo by stimulating osteoblast differentiation and suppressing osteoclast differentiation. Collectively, continuous overexpression of CLCF1 significantly improved both muscle and bone health, whereas a two-week administration exhibited limited effects in young mice.
CLCF1 deficiency in muscle impairs exercise-mediated improvements in muscle functions
Our in vivo and in vitro results highlighted the potential significance of CLCF1 in muscle and bone function. To definitively validate its functional role in muscle and bone, we sought to develop a muscle-specific genetic model. We intercrossed Clcf1 floxed mice with Mlc-Cre mice to generate skeletal muscle-specific Clcf1 knockout (Clcf1-skm KO) mice (Fig. 6A). Under sedentary conditions, no significant differences in Clcf1 mRNA expression were observed in muscle between Clcf1-skm KO and WTflx mice (Fig. S12A). However, in response to exercise, Clcf1-skm KO mice showed a significant reduction in Clcf1 expression specifically in muscle tissue, but not in other tissues, compared to WTflx mice (Figs. 6B and S12B). Exercise may trigger the activation of specific transcription factors or epigenetic modifications in skeletal muscles, leading to the induction of Clcf1 expression. Despite decreased muscle Clcf1 expression, plasma CLCF1 levels remained unchanged in both exercised and sedentary Clcf1-skm KO and WTflx mice (Fig. S12C). These findings imply that CLCF1 may be expressed in tissues other than skeletal muscle, including those we have not yet investigated, potentially contributing to the overall circulating CLCF1 pool. Sedentary Clcf1-skm KO male mice at 3 months of age exhibited no significant differences in body weight, grip strength, grid hanging performance, running capacity, or muscle weight compared to control WTflx male mice (Fig. S12D–H). We postulated that despite unaltered plasma CLCF1 levels, the autocrine effects of exercise-induced muscle-expressed CLCF1 on muscle function would be compromised in Clcf1-skm KO mice. Therefore, we performed a one-month high-resistance treadmill running and compared the phenotypes of WTflx and Clcf1-skm KO mice (Fig. 6A). Clcf1-skm KO mice exhibited significantly decreased muscle mass in both the TA and GA muscles compared to WTflx mice (Fig. 6C). Clcf1-skm KO mice exhibited reduced grip strength compared to WTflx mice, while grid hanging performance remained unaffected (Fig. 6D, E). Although Clcf1-skm KO mice showed a trend toward decreased running capacity, this difference did not reach statistical significance (Fig. 6F). Histological analysis revealed a significant decrease in the cross-sectional area of TA muscle fibers in Clcf1-skm KO mice compared to WTflx mice (Fig. 6G–I). However, analysis of isolated muscle tissue revealed no significant differences in the expression of genes associated with β-oxidation, glycolysis, or fiber type (Fig. S12I–K). Next, we analyzed the bones of exercised Clcf1-skm KO and WTflx mice. Micro-CT analysis showed there were no significant differences in trabecular bone volume, thickness, and number, cortical bone volume and thickness, and trabecular separation between WTflx and Clcf1-skm KO mice (Fig. 6J, K). Additionally, there were no significant differences in osteoclast and osteoblast numbers between the groups (Fig. 6L, M). The effect of skeletal muscle-specific Clcf1 deficiency on muscle and bone function is less pronounced than expected, even under exercise conditions. These findings suggest that exercise-induced CLCF1 expression may be upregulated in muscle fibers or other cell types within muscle tissue that are not targeted by Cre driven by the myofiber-specific promoters.
A Scheme showing the generation of skeletal muscle-specific CLCF1 knockout mice and the experimental plan of exercise (created in BioRender.com). We used 12-week-old male skm KO mice. B Relative mRNA expression levels of CLCF1 in TA muscle in WTflx and skm KO mice (n = 3; p = 0.0231). C Weight of dissected SOL, TA, EDL, and GA muscles in WTflx and skm KO mice (n = 5; biological replicates, p = 0.0271, 0.039). D Grip strength test (n = 5; p = 0.0026). E Grid hanging test (n = 5; p = 0.9296). F Treadmill running performance test in mice: running curves, distance run, and time to exhaustion (n = 5). G Immunostaining for myofiber boundaries using laminin in TA muscles of WTflx and skm KO mice. Scale bar, 50 μm. H, I Quantification of muscle fiber size and distribution in TA muscles: H mean muscle fiber area (WT, n = 4; skm KO, n = 3; biological replicates, p = 0.006), I frequency distribution of fiber area (WT, n = 4; skm KO, n = 3; biological replicates, p = 0.027, 0.045, 0.031). J Representative µCT images of the trabecular and cortical compartments of distal femoral metaphyseal regions of WTflx and skm KO mice. Scale bar, 1 mm. K Measurement of trabecular bone volume/total volume (BV/TV), trabecular thickness (TbTh), trabecular separation (TbSp), trabecular number (TbN), cortical bone volume/total volume (Corti BV/TV), and cortical thickness (Corti Th) (n = 5). L TRAP, hematoxylin/eosin (H&E), and Runx2 staining of a histological section of proximal tibiae. Scale bar, 100 μm. M Osteoclast number per bone surface and osteoblast number per bone surface were assessed (n = 5). Data are presented as means ± s.e.m.; two-tailed unpaired Student’s t-test (B–E, H–M); Log-rank (Mantel–Cox) test for running curve (F). *p < 0.05, **p < 0.005 and ***p < 0.001. Source data are provided as a Source Data file.
eCNTFR blocks the beneficial effect of high-resistance running in mice
Based on the partial effects of muscle-specific Clcf1 knockout, we next investigated alternative strategies to inhibit exercise-induced circulating CLCF1 activity. Building upon previous work, we have engineered a high-affinity soluble receptor, eCNTFR, which specifically binds to CLCF1 but blocks its interaction with gp130 and LIFR18 (Fig. S13A, B). eCNTFR pretreatment significantly decreased CLCF1-induced phosphorylation of STAT3, AKT, and ERK1/2 in C2C12 cells, whereas WT soluble CNTFR had no effect on CLCF1-mediated signaling (Fig. S13C, D). Additionally, eCNTFR pretreatment mitigated the effects of CLCF1 on basal, ATP-linked, and maximal mitochondrial respiration in C2C12 myotubes (Fig. S13E). Furthermore, eCNTFR injection efficiently reversed the anti-osteoclastogenic and pro-osteoblastogenic effects of CLCF1 (Fig. S13F, G). To evaluate the effect of blocking CLCF1 signaling in vivo, three-month-old male mice received vehicle or eCNTFR injections during one month of high-intensity treadmill running (Fig. 7A). After 4 weeks, there was no significant difference in body, heart, and epididymal white adipose tissues (WAT) weights among the four groups (Fig. S14A–C). There was no significant difference in muscle weight after 4 weeks of exercise (Fig. 7B). Although exercise enhanced grip strength, hanging time, and running performance compared with that in the control group, eCNTFR injection significantly abolished these exercise-induced improvements (Fig. 7C–G). Additionally, histological analysis showed that eCNTFR abolished the exercise-induced increase in muscle fiber size (Fig. S14D–G). To elucidate the underlying molecular mechanisms, we performed transcriptome analysis of the muscle tissue. eCNTFR injection significantly abolished exercise-induced upregulation of genes associated with FAO (Cpt1b and Hadha) and glycolysis (Hk1, Pkm2, and Pdk4) in muscle tissues (Fig. 7H, I). Treatment with eCNTFR also affected exercise-induced upregulation of genes related to ETC components and fiber type (Fig. S14H). Importantly, eCNTFR pretreatment significantly downregulated genes involved in ETC function (Atp5b and Ndufv2), glycolysis (Hk2), and FAO (Acaa2, Acad1, Acadm, Acox1, and Acsl1) even in non-exercised mice. Collectively, these results indicate that CLCF1 plays a critical role in mediating the beneficial effects of exercise on muscle function and metabolism.
A Experimental schemes for eCNTFR supplementation protocols during exercise (n = 8; created in BioRender.com). We used 12-week-old male mice. B Weight of dissected TA, GA, SOL, and EDL muscles (n = 8; p = 0.0674, 0.0754). C Grip strength test (n = 8; p = 0.0009, 0.0205). D Grid hanging test (n = 8; p = 0.0078, 0.0166). E–G Treadmill running performance test in mice: E running curves (p = 0.0025, 0.0285), F distance run (n = 7; p = 0.0014, 0.0172), and G time to exhaustion (n = 7; p = 0.0014, 0.0172). H, I mRNA expression of genes involved in fatty acid oxidation (n = 5; p = 1.21e-2, 4.89e-2, 2.3e-3, 5.0e-3, 3.0e-4, 4.0e-4, 1.0e-3, 1.2e-2, 5.0e-5, 1.5e-2, 1.4e-3, 8.0e-3, 8.0e-4) (H) and glycolysis (I) in TA muscles (n = 5, p = 8.2e-3, 8.8e-3, 1.71e-2, 4.0e-4, 6.5e-3, 2.12e-2). J Representative µCT images of the trabecular and cortical compartments of distal femoral metaphyseal regions of vehicle or eCNTFR-supplemented mice during exercise. Scale bar, 1 mm. K Measurement of trabecular bone volume/total volume (BV/TV, p = 3.0e-9, 5.0e-5), trabecular thickness (TbTh), trabecular separation (TbSp, p = 1.0e-7, 0.007), trabecular number (TbN, p = 1.0e-8, 0.0001), cortical bone volume/total volume (Corti BV/TV, p = 0.001, 0.001), and cortical thickness (Corti Th, n = 5). L TRAP, hematoxylin/eosin (H&E), and Runx2 staining of a histological section of proximal tibiae. Scale bar, 100 μm. M Osteoclast number per bone surface and osteoblast number per bone surface were assessed (n = 6; p = 0.0326, 0.0281, 0.0021, 0.0101). Data are presented as means ± s.e.m.; one-way ANOVA with post hoc Tukey’s multiple comparison test (B–M); or Log-rank (Mantel–Cox) test (E). *p < 0.05, **p < 0.005 and ***p < 0.001. Source data are provided as a Source Data file.
Furthermore, we investigated the effects of eCNTFR-induced CLCF1 inhibition on exercise-associated benefits in the bone. eCNTFR injection significantly suppressed the exercise-induced increase in bone mass (Fig. 7J). Micro-CT analysis revealed eCNTFR injection significantly reversed exercise-induced increase in bone volume and trabecular number and partially counteracted exercise-induced decrease in trabecular separation (Fig. 7K). Histological analysis further supported the involvement of osteoclasts and osteoblasts in the observed bone phenotypic changes. eCNTFR administration significantly reversed exercise-induced decrease in osteoclast numbers and attenuated exercise-induced increase in osteoblast numbers (Fig. 7L, M). Collectively, these findings demonstrate that CLCF1 plays a crucial role in mediating the beneficial effects of exercise on bone mass and microarchitecture, likely through its influence on osteoclast and osteoblast activity.
Discussion
Skeletal muscle and bone mass decline with age, leading to functional limitations. At the molecular level, aging alters the expression of several genes encoding proteins involved in mitochondrial function, the ubiquitin-proteasome proteolytic pathway, stress responses, and hormone/cytokine/growth factor signaling in muscles. However, little is known about age-related alterations in the expression of myokine genes in skeletal muscles. In this study, we examined age- and exercise-related alterations in myokine expression in human skeletal muscles. Notably, circulating CLCF1 levels decline with age in both mice and humans, but are restored by resistance exercise training. Our finding suggests that CLCF1 is an exercise-responsive myokine with therapeutic potential for mitigating age-related muscle and bone loss. Moreover, plasma CLCF1 may serve as a biomarker for monitoring the efficacy of exercise interventions in the elderly.
During exercise, muscle contractions require energy derived from ATP breakdown. Skeletal muscles utilize various energy systems to produce ATP, including phosphocreatine (PCr), anaerobic, and aerobic systems. Although aerobic metabolism serves as the predominant energy source during exercise, it is also essential for low- to moderate-intensity endurance. In contrast, high-intensity exercise requires a rapid rate of ATP resynthesis, primarily fueled by the ATP-PCr system and anaerobic glycolysis41. Notably, older men display reduced capacity for anaerobic glycolysis, a key energy source, during high-intensity exercise37. Glycolysis and glycolytic capacity were significantly lower in senescent human myoblasts42. Similarly, aged rats exhibited decreased LDH activity and LDHA mRNA expression in their skeletal muscles43. Additionally, proteomic and transcriptomic profiling of the muscle of elderly men with metabolic syndrome revealed a marked downregulation in specific glycolytic enzymes, including isoforms of PYGM, TPI1, ENO1, ENO3, and PKM, which are strongly associated with age-related muscle decline44. Similarly, metabolomic analysis of skeletal muscles from young and old mice has previously revealed decreased glycolytic pathway activity in aged mice36. Our present study demonstrates that CLCF1 treatment effectively reverses these age-related decreases in glycolytic enzymes, particularly PKM2, while promoting anaerobic glycolysis in mouse muscle tissue (Fig. 3I, J). Notably, when CLCF1 was inhibited through eCNTFR administration, the exercise-induced increase in PKM2 mRNA levels was abolished in exercised mice (Fig. 7I). These findings align with research by Verbrugge et al., who identified PKM2 as a key glycolytic enzyme regulated by resistance exercise and demonstrated its crucial role in muscle cell hypertrophy through isoform expression changes in human skeletal muscle.
CLCF1 was found to enhance glucose uptake and glycolysis (Fig. 3G, J), suggesting a role in supporting the energy demands of glycolytic fast-twitch fibers during acute resistance exercise. Consistently, CLCF1 secretion increased in response to acute resistance exercise in both humans and rodents. In addition, CLCF1 promoted mitochondrial metabolism (Fig. 3A), and its circulating levels were elevated following repeated bouts of acute exercise during long-term training, including in older individuals (Fig. S1). These findings raise the possibility that, in addition to its effects on fast-twitch fibers, CLCF1 may also act on oxidative slow-twitch fibers, which are rich in mitochondria. Together, these data suggest that CLCF1 contributes to fiber-type-specific metabolic regulation, promoting glycolytic flux in fast-twitch fibers and mitochondrial oxidative metabolism in slow-twitch fibers. This dual action highlights the versatility of CLCF1 in coordinating muscle energy metabolism in response to different forms of exercise.
Research evidence indicates that the IL-6 family of cytokines exerts their activities in a complex manner. Although members of the IL-6 family of cytokines are similar in structure and intracellular signaling, they have distinct biological activities and different binding affinities for receptors45. Although CLCF1 shares low homology (~23%) with CNTF, it has a highly conserved site that binds to CNTFR. CLCF1 or CNTF binds to a CNTFR-α, which induces the recruitment of gp130/ LIFRβ for signaling activation. Notably, CLCF1 and CNTF differ in their receptor specificity. For example, CNTF activates cells through IL-6Rα, LIFR-β, and gp130, whereas CLCF1/CRLF1 does not activate cells expressing these receptors46,47. Unlike CNTF, CLCF1 forms a complex with CRLF1 or soluble ciliary neurotrophic factor receptor (sCNTFR), not only for its secretion from target cells but also for receptor activation40. Recent studies reported that recombinant CLCF1 stimulates target cells without forming complexes with CRLF1 or sCNTFR18,48. Nahlé et al. reported that CLCF1 did not require CNTFR to stimulate mesenchymal stem cells during osteoblast differentiation and that a CLCF1 mutant with disrupted CNTFR binding activated mesenchymal stem cells16. CLCF1/CRLF1 and CNTF also use sortilin as a co-receptor to facilitate signaling49. To elucidate the underlying signaling pathways, we examined the effects of siRNA-mediated targeting of each receptor subunit on CLCF1 activation. LIFR or gp130 knockdown inhibited CLCF1-mediated phosphorylation of STAT3, AKT, and ERK1/2, whereas CNTFR or sortilin knockdown had no effect (Fig. S15A, B). In contrast, CNTF signaling was abrogated by CNTFR-α knockdown in C2C12 cells (Fig. S15C). Collectively, these findings suggest that CLCF1 may engage an alternative receptor to mediate downstream signaling in skeletal muscle. Further work is required to define the receptor(s) through which CLCF1 acts in skeletal muscle.
CLCF1 is detected in a wide range of tissues50. Our findings demonstrate that CLCF1 is predominantly upregulated in skeletal muscle in response to exercise stimuli (Fig. 1G). Despite confirming a significant reduction in CLCF1 mRNA levels in the skeletal muscle of clcf1-skm KO mice compared to WT, we observed no significant differences in plasma CLCF1 levels (Figs. 6B and S12C). This unexpected finding suggests the possibility of CLCF1 expression in uncharacterized tissues or other cell types within muscle during exercise. Furthermore, the effect of skeletal muscle-specific Clcf1 deficiency on muscle and bone function was less pronounced than anticipated, even under exercise conditions. These data suggest that muscle-specific knockout may have only partially disrupted the autocrine action of CLCF1, implying that its local effects are predominantly mediated through an autocrine mechanism. This notion is further supported by results in Fig. 7, where blockade of exercise-induced CLCF1 activity using eCNTFR attenuated the beneficial effects of exercise on both muscle and bone.
CLCF1 mutations can cause a syndrome characterized by cold-induced sweating and skeletal abnormalities21, emphasizing a potential role for CLCF1 in bone health. Chen et al. observed a decrease in CLCF1 expression in peripheral blood mononuclear cells (PBMCs) from postmenopausal women with osteoporosis compared with that in healthy individuals51. However, the precise role of CLCF1 in bone formation remains unresolved, with some reports suggesting a potential inhibitory effect. For example, Nahlé et al. demonstrated that CLCF1-induced STAT3 phosphorylation and suppressed the expression of osteoblast-specific genes in stem cells16. In contrast, Yokota et al. reported that CLCF1 promoted bone formation by suppressing osteoclast differentiation through the activation of interferon signaling and repression of the nuclear factor-kB pathway30. CLCF1 did not have any adverse effects on osteoblast differentiation in vivo or in vitro; rather, it increased the expression of Runx2, a master transcription factor for osteoblast differentiation. Overall, these conflicting findings highlight the need for further studies on the role of CLCF1 in bone health. In the present study, both external treatment and retroviral-induced overexpression of CLCF1 significantly suppressed osteoclast precursor cell differentiation through CLCF1-mediated activation of STAT1, which negatively regulates osteoclast development by inhibiting c-Fos and NFATc152. Additionally, the inhibitory effect of CLCF1 on osteoclastogenesis was strongly correlated with STAT1 activation and c-Fos and NFATc1 suppression. Moreover, the ectopic bone formation assay suggests a positive role for CLCF1 in promoting bone formation. Furthermore, mice with disrupted STAT3 signaling in osteoblasts exhibited an osteoporotic phenotype due to decreased bone formation53. In the present study, CLCF1 treatment increased STAT3 phosphorylation in osteoblasts. Additionally, STAT3 downregulation reversed the CLCF1-mediated increase in nodule formation to normal levels. Collectively, these findings suggest that CLCF1 positively regulates osteoblast differentiation via a STAT3-dependent pathway.
Our results showed inconsistent effects of CLCF1 on bone formation across different experimental models. In both in vitro cell models and young mouse models, CLCF1 promoted osteoblast formation and inhibited osteoclast formation (Fig. 4). However, in aging mouse models, the effect of CLCF1 on osteoblast formation was less pronounced (Fig. 2T). The diminished pool of stem and progenitor cells in aging bone compromises the tissue’s capacity for self-renewal and repair54. The limited effect of CLCF1 on osteoblast differentiation may be attributed to the age-related decline in the number of osteoblast precursor cells and stem cells. Consequently, in aged mice, CLCF1 appears to have a more significant impact on inhibiting bone resorption than on stimulating bone formation.
Aging is associated with structural and functional deterioration of the neuromuscular junction (NMJ), leading to impaired neuromuscular transmission and reduced muscle strength55,56,57. Although accumulating evidence suggests that regular physical activity preserves or restores NMJ integrity in aged muscle58, the underlying mechanisms remain incompletely understood. CLCF1 and its secretion partner CRLF1 are co-expressed in developing muscle59, and genetic ablation of either gene results in perinatal lethality due to motor neuron deficits and impaired suckling behavior19,60. Previous reports have demonstrated that CLCF1 promotes motor neuron survival in embryonic models14,61. Exercise-induced upregulation of CLCF1, together with these earlier findings, raises the possibility that CLCF1 may contribute to muscle function in part through motor neuron support. However, further studies are required to determine how CLCF1 modulates motor neuron integrity and neuromuscular function.
While the preclinical findings provide new insights into the physiological role of CLCF1, several limitations should be acknowledged. In particular, plasma CLCF1 levels at baseline and after exercise were notably higher in Brazilian Jiu-Jitsu athletes compared to other cohorts (Fig. S1D). Although these participants underwent standardized testing procedures, including a four-week familiarization period to ensure acclimatization, the elevated levels may reflect long-term physiological adaptations to high-intensity training. However, a systematic evaluation of the relationship between exercise intensity and circulating CLCF1 levels is lacking, and limited access to blood samples across varied exercise regimens constrained further analysis. Therefore, findings from the human cohorts should be interpreted as exploratory. Future studies incorporating well-characterized participants across a range of training intensities and durations will be required to clarify the potential association between CLCF1 and exercise load in humans.
This study identifies CLCF1 as an exercise-induced myokine with potential roles in attenuating age-related decline in muscle and bone. We show that CLCF1 expression is reduced in aged mice and elderly individuals, but can be restored through resistance exercise. In skeletal muscle, CLCF1 enhances glucose uptake, glycolytic flux, and mitochondrial metabolism, while in bone, it promotes formation via STAT3 signaling and suppresses resorption through STAT1 activation. These findings suggest that CLCF1 contributes to the beneficial effects of exercise on musculoskeletal health and may represent a potential therapeutic target for age-related musculoskeletal conditions. Furthermore, circulating CLCF1 levels may serve as a biomarker for assessing exercise responsiveness in older populations.
Methods
Transcriptomic analysis for identification of exercise-induced myokines
To identify the exercise-induced myokines, transcriptomic data (GSE28422)13 were analyzed using ‘GEOquery v2.60.0’ and ‘LIMMA v3.48.3’ in the R environment (v4.2). We followed the GEOquery-LIMMA pipeline to obtain and normalize the expression data, followed by the identification of differentially expressed genes in age groups between before and after exercise (p-value < 0.05, |log2FoldChange| > 1.5). To examine the expression of secretory protein genes, a list of predicted secretory proteins was downloaded from The Human Protein Atlas (2933 genes as of May 2022) at https://www.proteinatlas.org/humanproteome/tissue/secretome, and their expression fold-change values and p-values were extracted from the differential expression analysis result between pre- and post-exercise samples obtained earlier. Volcano plots were generated by the ‘ggplot2 v3.3.6’ package in R.
Ethics
Human cohorts 1–4 were conducted as Basic Experimental Studies involving Humans (BESH), and participants were not enrolled in a clinical trial. All procedures were approved by the relevant institutional review boards and were conducted in accordance with the Declaration of Helsinki. Human cohort 5 consisted of previously collected samples from a published study62, which reported the primary outcomes and was not considered a clinical trial. Human studies 1, 3, 4, and 6 were approved by the Institutional Review Board (IRB) at Pusan National University (IRB protocol number PNU_2021_140_HR). Human study 2 was approved by the IRB at Inha University (170621-1AC). Human study 5 was approved by the IRB at Seoul National University (SNUIRB no.1305/001-009). Written informed consent was obtained from all participants.
Human cohorts
Human study 1 (Young adults_acute resistance exercise): Ten young, active males (age 25.33 ± 1.837 years; height 178.4 ± 1.913 cm; weight 76.08 ± 2.983; BMI 23.84 ± 0.5952 kg/m2) males recruited to partake in this study. Participants were compensated with a monthly gym membership worth 45,000 won. Participants trained at a moderate intensity corresponding to 50–69% of their one-repetition maximum (1-RM), which is the heaviest weight they could lift for one repetition in this exercise. The exercise program is described in Table S1. Participants fasted for at least 8 h prior to exercise. Blood samples were collected before exercise and 1 h post-exercise.
Human study 2 (Young Jiu-Jitsu athletes_High-intensity interval exercise): Participants were selected based on the following criteria: a minimum of 4 years of Jiu-Jitsu training, training at least 4 days per week, a minimum of 8 h of training per week, and a history of placing in national Jiu-Jitsu competitions. Participants were compensated 200,000 won over the course of the study. Blood samples were collected from young Jiu-Jitsu athletes (age: 28.25 ± 3.62; height 176.63 ± 3.58 cm; weight 76.76 ± 8.62 kg; BMI 24.59 ± 2.48 kg/m²) before and immediately after completing the 4 rounds of the exercise program in a previous study by Park et al.63. Briefly, the high-intensity interval resistance exercise program, designed to mimic match conditions and induce exhaustion, consisted of six exercises, each performed for 1 min, followed by a 12-min rest period, and repeated for a total of 4 rounds. The exercises were: 1. Bar Cross Lateral Jump and Sprawl, 2. Bar hanging kimono holding, 3. 4-Step turtle control in the top position, 4. Gi grip jump over and roll back from the top position on the turtle, 5. Jump over the vaulting horse and tackle, 6. Rolling drill between the legs.
Human study 3 (Young adults_acute aerobic exercise): Ten young, active males (age 25.98 ± 1.74 years; height 175.9 ± 1.53 cm; weight 75.38 ± 2.53 kg; BMI 24.44 ± 0.98 kg/m²) participated in this study. Participants were compensated with a monthly gym membership worth 45,000 won. Participants fasted for a minimum of 8 h prior to exercise and then exercised on a treadmill for 60 min at a moderate intensity (heart rate between 40% and 59% of their maximum). The exercise program is described in Table S2. Blood was collected at rest before and one hour after exercise to evaluate plasma CLCF1 levels.
Human study 4 (Older adults_Chronic resistance training): Twenty-seven female older participants (age 72.32 ± 0.9484 years; height 155.7 ± 0.8637 cm; weight 60.04 ± 1.257 kg; BMI 24.79 ± 0.5447 kg/m²) participated in this study (details provided in Table S3). Participants were compensated with a monthly gym membership worth 45,000 won. Older participants exercised at 30~49% of 1-RM (light intensity) for one hour per day, 5 times per week for 16 weeks. Blood samples were collected from all participants the day after the final exercise session, following a minimum of 8 h of fasting. The exercise program is described in Table S4.
Human study 5 (Older adults_Chronic resistance band exercise): Blood samples were collected in a project investigating the effect of 12 weeks of elastic band-based high-speed power training on cognition and muscle function in elderly participants (n = 14, age 75.0 ± 3.46 years), as described in a study by Yoon et al.62. As a reward for their participation, participants were provided with exercise elastic bands and received personalized feedback on their exercise performance and blood test results. Briefly, the intensity was set by adjusting the length of the green elastic band (Hygenic Corporation, Akron, OH, USA). Participants engaged in a 12-week exercise program, completing two weekly 60-min sessions (Tuesday and Thursday). Each session included a 10-min warm-up, 40 min of elastic band training, and a 10-min cool-down period. Blood samples were collected before and after a 12-week exercise program in elderly participants. Following a minimum of 8 h of fasting, blood samples were collected from all participants the day after they completed the final exercise session.
Human study 6 (Plasma levels of CLCF1 in young and elderly subjects): To measure basal CLCF1 levels in young and elderly individuals, we utilized pre-exercise plasma samples from Human studies 1–5, along with additional plasma samples from 16 healthy young individuals and 10 healthy elderly individuals (age: 73.36 ± 4.69; height 153.57 ± 3.14 cm; weight 54.13 ± 4.12 kg; BMI 22.72 ± 1.92 kg/m²).
Animals
All animal experiments conducted in this study were approved by the Animal Care and Use Committee of the Korea Research Institute of Bioscience and Biotechnology (KRIBB). The mouse strains used in the experiments were of the C57BL/6J background. Young mice (3 months old), middle-aged mice (8 months old), and aged mice (18–24 months old) were obtained from the Laboratory Animal Resource Center (KRIBB). The mice were housed in cages maintained at 22–24 °C under a 12-h light/12-h dark cycle and were provided with free access to food and water. All mice were euthanized by CO2 inhalation followed by cervical dislocation. Serum and tissue samples were subsequently collected.
Generation of CLCF1-CRF1 TG mice and Clcf1-skeletal muscle-specific knockout mice
CLCF1 transgenic C57BL/6N mice were generated (Macrogen, Seoul, Korea). To facilitate efficient secretion, CRLF1 acted as a chaperone for CLCF140. A G4Sx3 linker was used to fuse CLCF1 with CRLF1. The cytomegalovirus (CMV) promoter was utilized to drive the expression of the Clcf1_G4Sx3_Crlf construct. Subsequently, the resulting plasmid, CMV_Clcf1_G4Sx3_Crlf_IRES_EmGFP, underwent digestion using NruI and PsiI enzymes to isolate the transgenic cassette. This expression cassette was purified and delivered through pronuclear injection into mouse zygotes with a C57BL/6N genetic background.
In order to create mice with skeletal muscle-specific Clcf1 deletion, mice harboring the conditional Clcf1 floxed allele were generated from Clcf1tm1a(EUCOMM)Wtsi ES cells obtained from EUCOMM. The floxed Clcf1 mice were kindly provided by the Korea Mouse Phenotyping Center (KMPC). They were bred with Flp recombinase mice to get homozygous Clcf1 floxed breeders. These breeders were crossed with Mlc-Cre mice to produce a strain of mice that lack Clcf1 specifically in their skeletal muscles.
Plasma sample assays ELISA
Blood samples were collected into tubes containing the anticoagulant EDTA (Greiner Bio-One, 450542) to prevent coagulation. The collected blood was then centrifuged at 1000 × g for 10 min at 4 °C to separate the plasma. After centrifugation, the plasma was carefully collected into new tubes to avoid contamination with cellular components and stored at −80 °C until further analysis. The concentration of CLCF1 in the plasma was measured using an enzyme-linked immunosorbent assay (ELISA) kit (USCN, SEC389Hu; Mybiosource, MBS762838) according to the manufacturer’s instructions. Mouse and human plasma samples were diluted 5-fold prior to performing the ELISA experiments. Samples were incubated at 37 °C for 1 h, followed by three washes with PBST (1 × PBS containing 0.05% Tween 20). The secondary antibody was diluted at 1:1000 in dilution buffer and incubated for 1 h. The plates were then washed three times with PBST, followed by incubation with TMB substrate solution at 37 °C for 20 min. The reaction was stopped by adding the Stop Solution. All samples and standards were assayed in duplicate, and absorbance was measured using a microplate reader.
Cell culture
The C2C12 mouse muscle cell line (ATCC, #CRL-1772) was maintained in a humidified incubator at 37 °C with 5% CO2 and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (100 U/mL, 100 μg/mL). To induce differentiation, C2C12 cells were grown in DMEM containing 2% FBS and 1% penicillin/streptomycin (Pen/Strep) until they formed myotubes. 3T3-L1 cells were differentiated in DMEM supplemented with 10% FBS, 1% Pen/Strep, and MDI (5 mM isobutylmethylxanthine (IBMX), 5 μg/mL insulin, and 1 μM dexamethasone) for 2 days following cell confluence. The cells were then maintained in medium containing 5 μg/mL insulin for an additional 2 days.
Primary osteoblast precursor cells were isolated from neonatal mouse calvariae using enzymatic digestion. The first set of cells following an initial treatment of calvariae for 10 min at 37 °C with 0.1% collagenase (Life Technologies, Carlsbad, CA) and 0.2% dispase II (Roche Diagnostics GmbH, Mannheim, Germany) were discarded. Primary osteoblast precursor cells were isolated from clavariae by sequential enzymatic digestion with collagenase for four 10-min periods. Osteoblast differentiation was then induced using osteogenic medium (OGM) containing 100 ng/mL BMP2, 50 μg/mL ascorbic acid, and 100 mM β-glycerophosphate for 3 or 6 days. ALP activity analysis was performed on day 3 of osteogenic differentiation. The following lysis of cells using an osteoblast lysis buffer [50 mM Tris-HCl (pH 7.4), 1% Triton X-100, 150 mM NaCl, and 1 mM EDTA], cell lysates were incubated with p-nitrophenyl phosphate substrate (Sigma-Aldrich, St Louis, MO) and ALP activity was determined by measuring the absorbance at 405 nm using a spectrophotometer. The alizarin red staining was performed after 6 days of osteogenic differentiation. The cultured cells were fixed with 70% ethanol and treated with a 40 mM alizarin red (pH 4.2) for 10 min. The stained culture was visualized with a CanoScan 9900F (Canon Inc., Japan), and then alizarin red was extracted with 10% acetic acid to quantify the substrate calcification. The concentration of alizarin red was determined by measuring the absorbance at 405 nm using a spectrophotometer.
Bone marrow cells were isolated from the long bones of ICR mice by flushing with α-MEM and incubated with a Red Blood Cell Lysis Buffer (Sigma-Aldrich, St. Louis, MO, USA). Bone marrow cells were cultured in α-MEM containing 10% FBS and antibiotics in the presence of M-CSF (30 ng/mL) for 3 days. Adherent cells (osteoclast precursors) were cultured in the presence of M-CSF and RANKL for 3 days. The cultured cells were fixed with 3.7% formalin and treated with TRAP staining solution (Sigma-Aldrich) for 10 min. The stained cells were visualized with a ProgRes CFscan camera (Jenoptik, Jena, Germany) and ProgRes Capture Pro software (Jenoptik). TRAP-positive multinucleated cells containing more than three nuclei were counted as osteoclasts.
Electrical pulse stimulation (EPS)
C2C12 myoblasts were seeded in six-well plates and differentiated into myotubes by culturing in differentiation medium (DMEM supplemented with 2% FBS) for 3 days. Following the differentiation period, the myotubes were subjected to electrical pulse stimulation (EPS) using the C-PACE EP culture pacer system (IonOptix, Dublin, Ireland) in a humidified incubator maintained at 37 °C with 5% CO2. The EPS protocol consisted of monophasic square wave pulses with a pulse duration of 2 milliseconds (ms) delivered at a frequency of 1 Hz. The voltage applied was 11.5 volts (V). Prior to the initiation of stimulation, cells were allowed to equilibrate in the stimulation setup for 1 h to recover from handling stress. Electrical stimulation was then applied continuously for 24 h under the specified conditions.
All stimulation parameters, including frequency, pulse width, and voltage, were optimized based on previous studies to mimic in vivo muscle contraction and ensure viability of C2C12 myotubes during prolonged stimulation.
Treadmill exercise
Mice underwent a gradual acclimatization process over two days, starting with a low incline and slow speed to familiarize them with the exercise equipment. Over the next two days, the incline was moderately increased while progressively extending the session duration. From day 5 onwards, the incline was further increased, and speed was slightly adjusted based on the mice’s tolerance. The objective was to push both young and aged mice near exhaustion without causing complete physical collapse. After exercise adaptation, all exercise bouts were performed at 70% of maximal speed on an incline greater than 5°. While young mice could easily maintain a speed of 24 m/min on a 10° incline, aged mice could only manage a speed of 15–18 m/min on a 5° incline. Each exercise bout was maintained for a total duration of 1 h. High-resistance running was defined as maintaining a speed of 24 m/min on a 10° incline. The chronic training protocol involved daily exercise sessions conducted over 30 consecutive days. Blood samples were collected from the mice before the exercise and immediately after the final exercise session.
Mitochondrial DNA contents
Genomic DNA was isolated from differentiated C2C12 myotubes using the Quick-DNA Miniprep Plus Kit (Zymo Research, D4068) according to the manufacturer’s instructions. Mitochondrial DNA (mtDNA) content was assessed by measuring the relative abundance of the mitochondrial gene ND1 to the nuclear gene HK2 using quantitative real-time PCR (qRT-PCR). The relative mtDNA content was calculated using the ΔCt method, where ΔCt = Ct(HK2) − Ct(ND1). Lower ΔCt values indicate a higher mitochondrial-to-nuclear DNA ratio. Primer sequences used for ND1 and HK2 are provided in Supplementary Table S6.
Glucose uptake and free fatty acid uptake assay
Glucose uptake was measured in vitro using differentiated C2C12 myotubes and 3T3-L1 adipocytes. After full differentiation, cells were serum-starved for 4 h in glucose-free DMEM prior to treatment. For stimulation of glucose uptake, cells were incubated with 1 μM insulin for 20 min at 37 °C. Following treatment, cells were washed with PBS, lysed, and analyzed using the Glucose Uptake Assay Kit (Abcam, ab136955) according to the manufacturer’s instructions. Absorbance was measured using a microplate reader at the recommended wavelength, and relative glucose uptake was quantified by interpolation from a standard curve. To measure free fatty acid (FFA) uptake, differentiated C2C12 myotubes and 3T3-L1 adipocytes were cultured in 96-well plates and treated with recombinant CLCF1 (100 ng/mL) for 24 h. FFA uptake was then assessed using the Free Fatty Acid Uptake Assay Kit (Abcam, ab176768), according to the manufacturer’s instructions. Briefly, after CLCF1 treatment, cells were incubated with a fluorescent fatty acid substrate provided in the kit, and uptake was measured using a fluorescence microplate reader at Ex/Em = 485/535 nm. Background signal was subtracted using control wells containing no cells, and relative fluorescence units (RFU) were normalized to total protein concentration per well. All experiments were performed in at least five independent biological replicates.
Glucose tolerance and insulin tolerance test
For glucose tolerance tests (GTT), mice were fasted overnight (10–12 h) before receiving an intraperitoneal injection of glucose at a dose of 2 g/kg body weight. Blood glucose levels were measured at 0, 15, 30, 60, 90, and 120 min using the ACCU-CHEK Performa test strip. Similarly, for insulin tolerance tests (ITT), mice were fasted for 4 h before receiving an intraperitoneal injection of insulin at a dose of 0.75 U/kg body weight. Blood glucose levels were measured at the same time points (0, 15, 30, 60, 90, and 120 min) using ACCU-CHEK Performa test strips. For both tests, blood samples were collected from the tail vein at the indicated time points.
Western blotting
Cells and tissues were lysed with RIPA buffer containing 0.1 mM Na3VO4, 1 mM NaF, 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (AEBSF), and 5 mg/ml aprotinin (Sigma-Aldrich). Total proteins were quantified using the BCA (bicinchoninic acid) assay kit and Bradford assay (Thermo Fisher, 23227). Protein samples were loaded on SDS-PAGE, transferred to polyvinylidene fluoride (PVDF) membranes and nitrocellulose (NC) membranes. The membranes were blocked for 1 h at RT with 5% skim milk or 2% BSA in Tris-buffered saline (TBS). The membrane was incubated with the respective primary antibodies at 4 °C overnight. All listed primary antibodies were used 1:1000. After washing with TBST, membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5000–1:10,000) for 1 h at RT. The image was detected using the ChemiDoc (Thermo Fisher, iBright CL1500). All primary antibodies used in this study are listed in Supplementary Table S5, along with their sources and catalog numbers.
Retroviral gene transduction
Retroviral vectors were transfected into the packaging cell line, Plat E, using FuGENE 6 (Promega, Madison, WI) according to the manufacturer’s instructions. Target cells were plated on 48-well or 6-well plates 24 h prior to infection. Viral supernatants, collected from the culture medium 48 h after transfection, were then incubated with the target cells for 6 h in the presence of 10 μg/mL polybrene (Sigma-Aldrich, St Louis, MO, USA).
Immunocytochemistry
C2C12 myoblasts were seeded on 24-well plates and differentiated into myotubes. Myotubes were rinsed in PBS, fixed in 4% paraformaldehyde for 15 min at room temperature, and washed three times with PBS and permeabilized in 0.25% Triton X-100 in PBS for 10 min. The myotubes were blocked with 1% BSA in PBS containing 0.1% Tween 20 for 30 min and incubated with anti-MYH (sc-376157) in blocking buffer overnight at 4 °C, followed by Alexa Fluor 488 (#A-21121, Thermo Fisher) with 1% BSA in PBS for 1 h at room temperature. And the myotubes were incubated with Vectashield with DAPI mounting medium (VECTOR Laboratories) for 1 min in the dark. Images were captured using a Nikon Eclipse Ti-U inverted microscope and Nikon DS-Ri2 camera using NIS-Elements software. The short-axis diameters of myotubes were analyzed by Image J software.
Immunohistology
Muscles were mounted in optimal cutting temperature medium (O.C.T, Tissue-Tek) at resting length and frozen in dry ice and stored at −80 °C. Muscle sections (7 μm) were cut from the mid at −20 °C on a cryostat (Leica, CM1950) and collected on Micro Slides (MUTO, Japan) and stored at −80 °C. For immunofluorescence staining, serial sections were air-dried for 30 min and washed three times with PBS. Sections were blocked and permeabilized in PBS and 0.4% Triton X-100 for 30 min before being incubated for 2 h at room temperature in a primary antibody solution containing laminin (L9393, Sigma-Aldrich) and 10% goat serum (ab7481, Abcam) followed by Alexa555 (A32732, Invitrogen), Alexa555 (A28180, Invitrogen) in PBS containing 10% goat serum. Sections were washed three times in PBS and mounted with Vectashield with DAPI mounting medium (VECTOR Laboratories). Images were captured using a Nikon Eclipse Ti-U inverted microscope and Nikon DS-Ri2 camera using NIS-Elements software.
Quantitative RT-PCR (qPCR)
Total RNA was isolated from tissues and cells using RiboEx reagent (GeneAll Biotechnology Co., South Korea). cDNA synthesis was performed using the iScript cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR analysis was performed using StepOnePlus (Applied Biosystems) with a 20 μl reaction volume containing cDNA, primers, and SYBR Master Mix (Applied Biosystems). The sequences of the primers used in this study are provided in Supporting Information, Table S6.
CLCF1 supplementation
3-month-old and 20-month-old C57BL6 mice were injected intraperitoneally with either recombinant CLCF1 (Abcam, ab50021 or Enzynomics, C045) or PBS (vehicle control) at a dose of 0.1 or 0.5 mg/kg body weight per day for 14 consecutive days.
eCNTFR production and supplementation
We used an engineered CNTFR (eCNTFR)18 to achieve effective blockade of CLCF1. This blockade requires both increased binding of the decoy to CLCF1 and decreased binding of CLCF1 to gp130 and LIFR. The eCNTFR-Fc recombinant protein, containing eight consensus mutations (R110Q, T174P, S237F, I287F, Y177H, K178N, T268A, D269A), was synthesized by Y-Biologics (Y-Biologics, Deajeon, South Korea). Mice were treated with eCNTFR-Fc (10 mg/kg) or PBS (vehicle) by i.p injection for 1 month, three times per week.
Production of CLCF1/CRLF1 protein complex
The human Cardiotrophin-like cytokine factor 1 (hCLCF1, Uniprot Q9UBD9) and human Cytokine receptor-like factor 1 (hCRLF1, Uniprot O75462) genes were amplified by PCR (Sogent, SPX16-R250). Amplicons were digested with BamHI (New England Biolabs, R0136) / XhoI (New England Biolabs, R0146) and inserted into the pcDNA3.4 vector (Thermo Fisher Scientific, A14697). A polyhistidine tag was placed at the C-terminus of CLCF1 for affinity chromatography. For transient expression of CLCF1/CRLF1 protein complex, Expi293FTM cells (Thermo Fisher Scientific, A14527) were cultured in Expi293TM expression medium (Thermo Fisher Scientific, A1435102). The cells were maintained in a humidified chamber at 37 °C with 8% CO2. ExpiFectamineTM 293 transfection kit (Thermo Fisher Scientific, A14525) was used following the manufacturer’s instructions for efficient transfection of Expi293FTM cells with plasmids. Briefly, the cells were diluted at a density of 1.7 × 106 cells/ml in 300 ml of fresh media in 1 L Erlenmeyer shake flask (Corning, 7513). On the following day, DNA plasmids encoding hCLCF1 and hCRLF1 genes and ExpiFectamineTM 293 were separately mixed in Opti-MEMTM (Thermo Fisher Scientific, 31985070) and incubated at room temperature for 5 min. Then, the ExpiFectamineTM 293 and DNA mixtures were combined and incubated for another 20 min at room temperature. Finally, the mixture was added to flasks containing the cells and incubated in a humidified chamber at 37 °C with 8% CO2. After 18 h, a cocktail of Enhancer I and II was added to the flask. The cells were then incubated for 72~96 h, and the culture media were collected. Recombinant CLCF1/CRLF1 protein complex was purified by affinity and size exclusion chromatography using a fast protein liquid chromatography machine (FPCL) (Cytiva, ÄKTA Go system). The flited culture media was applied at a flow rate of 5 ml/min to the HisTrap Excel column (Cytiva, 17371205) previously equilibrated with 20 mM imidazole, 500 mM NaCl, and 20 mM Na2HPO4 at pH 7.4. Unbound proteins were washed away with 50 ml equilibration buffer. Bound fractions were eluted with 75 mL elution buffer containing a 20-to-500 mM imidazole gradient in 20 mM Na2HPO4 and 500 mM NaCl at pH 7.4 for 15 min. The protein complex was collected, pooled, and analyzed by SDS-PAGE. The eluted proteins were polished by SEC on an FPLC machine using a HiLoad 16/600 Superdex 200 pg (Cytiva, 28989335) previously equilibrated with PBS (pH 7.4) at a flow rate of 0.5 ml/min. Finally, Fractions containing CLCF1/CRLF protein complex were collected, pooled, and analyzed by SDS-PAGE and immunoblot analysis.
Glycolysis measurement and oxygen consumption rate
Oxygen consumption rates of C2C12 myotube cells, seeded at a density of X cells/well in a 96-well cell culture plate, were recorded on an XF96 Extracellular Flux Analyzer using the XF Cell Mito Stress Test Kit (Seahorse Bioscience, North Billerica, MA) according to the manufacturer’s protocol. Following seeding, cells were washed and pre-incubated in XF DMEM media (pH 7.4) for 1 h prior to the assay. The XF Cell Mito Stress Test was then performed according to the manufacturer’s protocol. Oligomycin (1 μM), FCCP (2 μM), and rotenone & antimycin A (0.5 μM each Rot/Anti) were added as indicated. All measured values were normalized to total protein content, which was quantified using the BCA protein assay.
siRNA transfection
C2C12 cells were seeded in 24-well plates with 1 × 105 cells per well and then differentiated into myotubes for 4 days using a differentiation medium (2% FBS DMEM). At 3 days during differentiation, myotube was transiently transfected with siRNA control (Dhamacon, ON-TARGET plus siRNA) or siRNA Cntfr, Il6st (gp130), and Lifr using Lipofectamin RNAiMAX reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s protocols. Primary osteoblast precursor cells or osteoclast precursor cells were transfected with siRNA control, siRNA Stat1, or siRNA Stat3 (Dhamacon, ON-TARGET plus siRNA) using Lipofectamine RNAiMAX (Thermo Fisher Scientific, USA), according to the manufacturer’s instructions.
Skeletal muscle function exploration
Time and distance to exhaustion
After five consecutive days of acclimation, running performance was tested in mice by measuring their ability to run until exhaustion. Mice were exercised on a treadmill at 10 m/min for 10 min, and the speed was increased every 2 min by 2 m/min until 24 m/min. Mice were considered to be exhausted when mice remained on the electric shock grid for more than 10 s. Time and distance were recorded and calculated.
Strength by grip test
Mice were allowed to grasp an angled grid with all four limbs. They were then gently pulled backwards until they lost their grip, for a total of three attempts. The force (in Newtons) applied to the grid during each attempt was recorded. The average of these three measurements was then normalized to the body weight of each mouse.
Inverted grid test
Mice were placed in the center of a wire grid system, which was then inverted and held steadily 40 cm above a padded surface. The latency to fall, measured by the time the mice spent hanging onto the grid system, was then recorded.
Ex vivo isometric force and fatigue measurements
Intact TA muscles were isolated from the hindlimb of euthanized mice and mounted vertically between a force transducer (Model FT03, Glass Instruments, USA) in an organ bath with platinum electrodes and continuous perfusion with 95% O2 + 5% CO2-saturated Krebs–Ringer solution (118 mM NaCl, 4.75 mM KCl, 24.8 mM NaHCO3, 1.18 mM KH2PO4, 2.5 mM CaCl2 ∙ 2H20, 1.18 mM MgSO4, and 10 mM glucose). Optimal muscle stretch was determined by applying a single twitch at a supramaximal voltage of 100 V for 1 ms, and set at the length that generated maximal twitch force after 10 min of equilibration, TA muscles were tested with a range of stimulation frequencies (tetanic with increasing frequencies from 30 to 200 Hz every 500 ms, with 2-min recovery intervals between each frequency). The fatigue properties of the TA muscles were assessed by repeated stimulation at 1 Hz and 100 V for 10 min. All experiments were conducted at 25 °C. Data acquisition and analysis were performed using LabChart Pro Software (Version 8; AD Instruments, Pty Ltd.). At the end of each experiment, muscle length, diameter, and wet weight were measured.
RANKL-induced bone loss and ectopic bone formation
For the RANKL-induced bone loss mouse model64, 7-week-old male ICR mice were divided into three groups: PBS, RANKL with PBS, and RANKL with CLCF1. PBS or CLCF1 (0.2 mg/kg) was intraperitoneally injected for 4 consecutive days. From the second day, PBS, RANKL with PBS, or RANKL with CLCF1 was intraperitoneally administered for 3 consecutive days. For the ectopic bone formation model, a sponge soaked with BMP2 (1 µg) and PBS or a sponge soaked with BMP2 (1 µg) and CLCF1 (1 µg) was subcutaneously implanted on the left or right side, respectively, of 5-week-old male ICR mice. After four weeks, all sponges were collected from mice and then subjected to micro-computed tomography analysis.
Micro-computed tomography (μCT) analysis
Distal femurs were scanned with a SkyScan 1172 system (SkyScan, Kontich, Belgium) using the following parameters: 50 kV, 201 μA, 0.5 mm aluminum filter, and a resolution of 11 μm per pixel. Images were captured every 0.7° over a 180° angular range. The raw images were reconstructed into serial cross-sections using NRecon 1.4 (SkyScan) for analysis of femoral morphometric parameters. CTAn (SkyScan) and Ant 2.4 (SkyScan) software were used for further data analysis and 3D model visualization, respectively.
Histological analysis
Harvested tibias from mice were fixed with 4% paraformaldehyde overnight, followed by decalcification in 3.8% formaldehyde buffer containing 5.5% EDTA for 3 weeks at 4 °C. Decalcified tibias were dehydrated and embedded in paraffin blocks. The paraffin blocks were cut into 4-μm-thick longitudinal sections. The sectioned samples were deparaffinized using xylene and subjected to tartrate-resistant acid phosphatase (TRAP) staining to identify osteoclasts located below the growth plate and hematoxylin & eosin (H&E) and Runx2 (ab192256, Abcam) staining to identify osteoblasts located below the growth plate.
Statistical analysis
All data here are expressed as the mean + standard error of the mean (SEM). Statistical analyses were performed using Student’s t-test (two-tailed paired or unpaired), one-way ANOVA followed by the post hoc Bonferroni’s and Tukey’s test, or two-way ANOVA followed by the post hoc Bonferroni’s and Tukey’s test with GraphPad Prism 10 software (GraphPad Software). p-values less than 0.05 (* or #), 0.01 (** or ##) or 0.001 (*** or ###) were statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper. The dataset generated in this study was derived from a reanalysis of data available in the GEO database under accession code GSE28422. Additional datasets used and/or analyzed during the current study are available as the Supplementary Information files. Source data are provided with this paper.
References
Tagliaferri, C., Wittrant, Y., Davicco, M. J., Walrand, S. & Coxam, V. Muscle and bone, two interconnected tissues. Ageing Res. Rev. 21, 55–70 (2015).
Yu, X. et al. A pooled analysis of the association between sarcopenia and osteoporosis. Medicine 101, e31692 (2022).
Leal, L. G., Lopes, M. A. & Batista, M. L. Jr. Physical exercise-induced myokines and muscle-adipose tissue crosstalk: a review of current knowledge and the implications for health and metabolic diseases. Front. Physiol. 9, 1307 (2018).
Karsenty, G. & Mera, P. Molecular bases of the crosstalk between bone and muscle. Bone 115, 43–49 (2018).
Bostrom, P. et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481, 463–468 (2012).
Miyamoto-Mikami, E. et al. Endurance training-induced increase in circulating irisin levels is associated with reduction of abdominal visceral fat in middle-aged and older adults. PLoS ONE 10, e0120354 (2015).
Kim, H. J., So, B., Choi, M., Kang, D. & Song, W. Resistance exercise training increases the expression of irisin concomitant with improvement of muscle function in aging mice and humans. Exp. Gerontol. 70, 11–17 (2015).
Reza, M. M. et al. Irisin is a pro-myogenic factor that induces skeletal muscle hypertrophy and rescues denervation-induced atrophy. Nat. Commun. 8, 1104 (2017).
Colaianni, G. et al. The myokine irisin increases cortical bone mass. Proc. Natl. Acad. Sci. USA 112, 12157–12162 (2015).
Qiao, X. et al. Irisin promotes osteoblast proliferation and differentiation via activating the MAP kinase signaling pathways. Sci. Rep. 6, 18732 (2016).
Vinel, C. et al. The exerkine apelin reverses age-associated sarcopenia. Nat. Med. 24, 1360–1371 (2018).
Hang, K. et al. Apelin enhances the osteogenic differentiation of human bone marrow mesenchymal stem cells partly through Wnt/beta-catenin signaling pathway. Stem Cell Res. Ther. 10, 189 (2019).
Raue, U. et al. Transcriptome signature of resistance exercise adaptations: mixed muscle and fiber type specific profiles in young and old adults. J. Appl Physiol. 112, 1625–1636 (2012).
Senaldi, G. et al. Novel neurotrophin-1/B cell-stimulating factor-3: a cytokine of the IL-6 family. Proc. Natl. Acad. Sci. USA 96, 11458–11463 (1999).
Cognet, I. et al. Cardiotrophin-like cytokine labelling using Bir A biotin ligase: a sensitive tool to study receptor expression by immune and non-immune cells. J. Immunol. Methods 301, 53–65 (2005).
Nahle, S. et al. Cardiotrophin-like cytokine (CLCF1) modulates mesenchymal stem cell osteoblastic differentiation. J. Biol. Chem. 294, 11952–11959 (2019).
Senaldi, G. et al. Regulatory effects of novel neurotrophin-1/b cell-stimulating factor-3 (cardiotrophin-like cytokine) on B cell function. J. Immunol. 168, 5690–5698 (2002).
Kim, J. W. et al. Antitumor activity of an engineered decoy receptor targeting CLCF1-CNTFR signaling in lung adenocarcinoma. Nat. Med. 25, 1783–1795 (2019).
Alexander, W. S. et al. Suckling defect in mice lacking the soluble haemopoietin receptor NR6. Curr. Biol. 9, 605–608 (1999).
Forger, N. G. et al. Cardiotrophin-like cytokine/cytokine-like factor 1 is an essential trophic factor for lumbar and facial motoneurons in vivo. J. Neurosci. 23, 8854–8858 (2003).
Rousseau, F. et al. Inactivation of cardiotrophin-like cytokine, a second ligand for ciliary neurotrophic factor receptor, leads to cold-induced sweating syndrome in a patient. Proc. Natl. Acad. Sci. USA 103, 10068–10073 (2006).
Hahn, A. F. et al. Cold-induced sweating syndrome: CISS1 and CISS2: manifestations from infancy to adulthood. Four new cases. J. Neurol. Sci. 293, 68–75 (2010).
Pourteymour, S. et al. Global mRNA sequencing of human skeletal muscle: Search for novel exercise-regulated myokines. Mol. Metab. 6, 352–365 (2017).
Behnke, B. J. et al. Effects of aging and exercise training on skeletal muscle blood flow and resistance artery morphology. J. Appl Physiol. 113, 1699–1708 (2012).
Popov, D. V. et al. Contractile activity-specific transcriptome response to acute endurance exercise and training in human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 316, E605–E614 (2019).
Romero, S. A. et al. Evidence of a broad histamine footprint on the human exercise transcriptome. J. Physiol. 594, 5009–5023 (2016).
Toledo-Corral, C. M. & Banner, L. R. Early changes of LIFR and gp130 in sciatic nerve and muscle of diabetic mice. Acta Histochem. 114, 159–165 (2012).
Helgren, M. E. et al. Trophic effect of ciliary neurotrophic factor on denervated skeletal muscle. Cell 76, 493–504 (1994).
Guillet, C., Auguste, P., Mayo, W., Kreher, P. & Gascan, H. Ciliary neurotrophic factor is a regulator of muscular strength in aging. J. Neurosci. 19, 1257–1262 (1999).
Yokota, S. et al. Cardiotrophin Like Cytokine Factor 1 (CLCF1) alleviates bone loss in osteoporosis mouse models by suppressing osteoclast differentiation through activating interferon signaling and repressing the nuclear factor-kappaB signaling pathway. Bone 153, 116140 (2021).
Kim, J. W. et al. Engineering a potent receptor superagonist or antagonist from a novel IL-6 family cytokine ligand. Proc. Natl. Acad. Sci. USA 117, 14110–14118 (2020).
Sartori, R., Romanello, V. & Sandri, M. Mechanisms of muscle atrophy and hypertrophy: implications in health and disease. Nat. Commun. 12, 330 (2021).
Wegrzyn, J. et al. Function of mitochondrial Stat3 in cellular respiration. Science 323, 793–797 (2009).
Carbognin, E., Betto, R. M., Soriano, M. E., Smith, A. G. & Martello, G. Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J. 35, 618–634 (2016).
Knudsen, N. H. et al. Interleukin-13 drives metabolic conditioning of muscle to endurance exercise. Science 368, eaat3987 (2020).
Uchitomi, R. et al. Metabolomic analysis of skeletal muscle in aged mice. Sci. Rep. 9, 10425 (2019).
Lanza, I. R., Befroy, D. E. & Kent-Braun, J. A. Age-related changes in ATP-producing pathways in human skeletal muscle in vivo. J. Appl Physiol. 99, 1736–1744 (2005).
Zhang, F. et al. Cytokine Receptor-like Factor 1 (CRLF1) and its role in osteochondral repair. Cells 13, 757 (2024).
Johnson, R. W. et al. Glycoprotein130 (Gp130)/interleukin-6 (IL-6) signalling in osteoclasts promotes bone formation in periosteal and trabecular bone. Bone 81, 343–351 (2015).
Elson, G. C. et al. CLF associates with CLC to form a functional heteromeric ligand for the CNTF receptor complex. Nat. Neurosci. 3, 867–872 (2000).
Hargreaves, M. & Spriet, L. L. Skeletal muscle energy metabolism during exercise. Nat. Metab. 2, 817–828 (2020).
Rajabian, N. et al. Methionine adenosyltransferase2A inhibition restores metabolism to improve regenerative capacity and strength of aged skeletal muscle. Nat. Commun. 14, 886 (2023).
Washington, T. A., Healey, J. M., Thompson, R. W., Lowe, L. L. & Carson, J. A. Lactate dehydrogenase regulation in aged skeletal muscle: regulation by anabolic steroids and functional overload. Exp. Gerontol. 57, 66–74 (2014).
Gueugneau, M. et al. Muscle proteomic and transcriptomic profiling of healthy aging and metabolic syndrome in men. Int. J. Mol. Sci. 22, 4205 (2021).
Metcalfe, R. D., Putoczki, T. L. & Griffin, M. D. W. Structural understanding of interleukin 6 family cytokine signaling and targeted therapies: focus on interleukin 11. Front. Immunol. 11, 1424 (2020).
Tormo, A. J. et al. The cytokines cardiotrophin-like cytokine/cytokine-like factor-1 (CLC/CLF) and ciliary neurotrophic factor (CNTF) differ in their receptor specificities. Cytokine 60, 653–660 (2012).
Schuster, B. et al. Signaling of human ciliary neurotrophic factor (CNTF) revisited. The interleukin-6 receptor can serve as an alpha-receptor for CTNF. J. Biol. Chem. 278, 9528–9535 (2003).
Pasquin, S. et al. Cardiotrophin-Like Cytokine Factor 1 exhibits a myeloid-biased hematopoietic-stimulating function. Front. Immunol. 10, 2133 (2019).
Larsen, J. V. et al. Sortilin facilitates signaling of ciliary neurotrophic factor and related helical type 1 cytokines targeting the gp130/leukemia inhibitory factor receptor beta heterodimer. Mol. Cell Biol. 30, 4175–4187 (2010).
Elson, G. C. et al. Cytokine-like factor-1, a novel soluble protein, shares homology with members of the cytokine type I receptor family. J. Immunol. 161, 1371–1379 (1998).
Chen, X. et al. Association of cardiotrophin-like cytokine factor 1 levels in peripheral blood mononuclear cells with bone mineral density and osteoporosis in postmenopausal women. BMC Musculoskelet. Disord. 22, 62 (2021).
Furukawa, M. et al. IL-27 abrogates receptor activator of NF-kappa B ligand-mediated osteoclastogenesis of human granulocyte-macrophage colony-forming unit cells through STAT1-dependent inhibition of c-Fos. J. Immunol. 183, 2397–2406 (2009).
Itoh, S. et al. A critical role for interleukin-6 family-mediated Stat3 activation in osteoblast differentiation and bone formation. Bone 39, 505–512 (2006).
Kassem, M. & Marie, P. J. Senescence-associated intrinsic mechanisms of osteoblast dysfunctions. Aging Cell 10, 191–197 (2011).
Deschenes, M. R., Roby, M. A., Eason, M. K. & Harris, M. B. Remodeling of the neuromuscular junction precedes sarcopenia related alterations in myofibers. Exp. Gerontol. 45, 389–393 (2010).
Butikofer, L., Zurlinden, A., Bolliger, M. F., Kunz, B. & Sonderegger, P. Destabilization of the neuromuscular junction by proteolytic cleavage of agrin results in precocious sarcopenia. FASEB J. 25, 4378–4393 (2011).
Li, Y., Lee, Y. & Thompson, W. J. Changes in aging mouse neuromuscular junctions are explained by degeneration and regeneration of muscle fiber segments at the synapse. J. Neurosci. 31, 14910–14919 (2011).
Valdez, G. et al. Attenuation of age-related changes in mouse neuromuscular synapses by caloric restriction and exercise. Proc. Natl. Acad. Sci. USA 107, 14863–14868 (2010).
de Bovis, B. et al. clc is co-expressed with clf or cntfr in developing mouse muscles. Cell Commun. Signal. 3, 1 (2005).
Zou, X. et al. Neonatal death in mice lacking cardiotrophin-like cytokine is associated with multifocal neuronal hypoplasia. Vet. Pathol. 46, 514–519 (2009).
Plun-Favreau, H. et al. The ciliary neurotrophic factor receptor alpha component induces the secretion of and is required for functional responses to cardiotrophin-like cytokine. EMBO J. 20, 1692–1703 (2001).
Yoon, D. H. et al. Effect of elastic band-based high-speed power training on cognitive function, physical performance and muscle strength in older women with mild cognitive impairment. Geriatr. Gerontol. Int. 17, 765–772 (2017).
Park, J. H. et al. Physical performance and inflammation responses during and after the Jiu-Jitsu stimulated match according to recovery treatment methods. Exerc. Sci. 29, 214–224 (2020).
Kim, J. H. et al. Lhx2 regulates bone remodeling in mice by modulating RANKL signaling in osteoclasts. Cell Death Differ. 21, 1613–1621 (2014).
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) Grants funded by the Korean Government (2022R1A2C1006418 and 2019R1A5A2027521), the Science and Technology Project that Opens the Future of the Region (2021-DD-UP-0380-03-203) of the Korea Innovation Foundation (INNOPOLIS) funded by the South Korean government (Ministry of Science and ICT), the National Research Council of Science & Technology (NST) grant by the Korean government (MSIT) (CRC22011−200), and the KRIBB initiative program.
Author information
Authors and Affiliations
Contributions
J.S.K., J.H.K., and M.J.K. designed and performed the experiments. B.M. and C.K. performed bioinformatic analysis. S-M.L., G-Y.G., J-W.K., S.K., and J.Y.K., performed the experiments. S-W.C., W.S., H.Y.M., S.G.C., and D-H.P. provided human plasma samples. J.H.P. conducted protein production and activity assays. K-P.L. and E-S.K. provided recommendations for the experiments and interpreted data. N.K., K-S.K., and Y.R.Y. conceptualized the study, designed experiments, and interpreted data. J.S.K., J.H.K., and Y.R.Y. wrote the manuscript. Y.R.Y. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Kohjiro Ueki and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kang, J.S., Kim, J.H., Kim, M.J. et al. Exercise-induced CLCF1 attenuates age-related muscle and bone decline in mice. Nat Commun 16, 4743 (2025). https://doi.org/10.1038/s41467-025-59959-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-59959-w