Abstract
Toxoplasma gondii, an apicomplexan parasite and agent of the human disease toxoplasmosis, possesses a non-photosynthetic relic plastid, named the apicoplast. Thought to be evolved from a red algal plastid, the apicoplast houses major metabolic pathways, such as heme, isoprenoid and lipid synthesis, crucial for parasite survival, and thus considered attractive drug targets. However, despite similarities with plant chloroplast lipid synthesis pathways, the apicoplast lacks canonical plant/chloroplast lipid transporters and so metabolite import/export is at present, poorly characterised. Here we identify TgFLP12, a newly identified P5-ATPase transporter localised to the Toxoplasma apicoplast. TgFLP12 is found in the SAR (Stramenopile-Alveolata-Rhizaria) supergroup (to which belong Apicomplexa parasites and chromerids) but absent in higher plants. Disruption of TgFLP12 causes major defects on apicoplast morphology. Lipidomic analyses and stable isotope labelling reveal a unique accumulation of C14:0 in the apicoplast, which is then lacking in most major lipid classes subsequently synthesized in the ER. Successful complementation of a yeast mutant deficient in fatty acid transport with TgFLP12 validates TgFLP12 as a fatty acid transporter. Overall, we identify a potentially important drug target: the apicoplast fatty acid exporter, specific to Apicomplexa which unexpectedly also highlights Toxoplasma’s utility as a model organism for investigating algal biology.
Similar content being viewed by others
Introduction
The phylum Apicomplexa includes unicellular infectious pathogens responsible for major human diseases with heavy health and economic burdens (www.who.org). it includes, Toxoplasma gondii, the agent of human toxoplasmosis, a global infectious chronic disease that affects about a third of the world population1. To maintain intracellular development and a high asexual division rate, Apicomplexa need large amounts of lipids, which they obtain through a combination of host cell scavenging, and de novo synthesis via the parasite’s own metabolic pathways. Fatty acids (FAs) are the pivotal hydrophobic lipid buildings block at the center of all scavenging, synthesis, and trafficking pathways2,3,4,5. The parasite is capable of sensing and metabolically adapting the activity of each FA acquisition pathway (host scavenging vs de novo synthesis) upon the host’s nutritional content6,7.
The current consensus is based on the following:
On the one hand, the parasite constantly and massively scavenges FAs directly from the host cell and its external environment4,5,6,7. This constant flux of host FAs is channeled toward parasite lipids storages, i.e., lipid droplets (LD), by TgLIPIN, in the form of triacylglycerol (TAG). LDs are then timely mobilized, specifically during parasite division, to allow parasite propagation and avoid lipotoxic/lethal accumulation of FAs in the parasite6.
On the other hand, most Apicomplexa harbor a relict non-photosynthetic plastid named the apicoplast, which was acquired by the secondary endosymbiosis of a red algal ancestor8,9,10. The apicoplast harbors a prokaryotic type II fatty acid synthesis pathway (FASII) that is essential for parasite survival depending on the parasite life stage and host nutrient content11,12,13,14. Specifically, the apicoplast FASII acts as the second pivotal provider of FAs for the parasite; it plays critical roles when host lipid content varies and becomes lower: the parasite senses this and up-regulates apicoplast FASII activity so it becomes an essential provider of FAs for membrane biogenesis and parasite survival14,15,16,17.
In photosynthetic plastids, the essential FASII provides FAs for the de novo synthesis of galactolipids that are essential for chloroplast biogenesis and photosynthesis18. However, in Apicomplexa, the function of the apicoplast FASII has been repurposed to fit the obligate intracellular lifestyle of obligate intracellular parasites14. We, and others, showed that instead of fueling plastid galactolipid synthesis, which is absent in Apicomplexa, the apicoplast FASII provides FAs for the bulk synthesis of all parasite phospholipids (PL), which then allows membrane biogenesis and parasite survival14. More precisely, the parasite uses glycolytic intermediates to generate short FAs lauric acid (C12:0), myristic acid (C14:0), and palmitic acid (C16:0) via the apicoplast FASII. These FAs are (i) used via a plant-like acyltransferase TgATS1 (or G3PAT in Plasmodium spp.) to form lysophosphatidic acid (LPA) This LPA serves as a central lipid precursor that is exported to the ER and combined with host scavenged FAs, forming an “obligate FA-patchwork PA”, as a central precursor for bulk phospholipid synthesis and parasite membrane biogenesis. FAs are also (ii) directly exported from the apicoplast to the ER, for further elongation and later incorporation for bulk phospholipid synthesis as well. Hence, FA -and LPA- must be exported from the apicoplast to reach the ER for further elongation and incorporation into PL.
The Toxoplasma lipidome is particularly enriched in myristic acid, between 10–15% of total FAs, compared to the host (human foreskin fibroblast (HFF) or serum (fetal bovine serum (FBS)) for which myristic acid represents around 3% of the total FAs19. The parasite synthesizes FA chains via its apicoplast FASII pathway, in which a core protein is the acyl carrier protein (ACP). ACP structure analysis in Plasmodium reveals that its hydrophobic groove that contains the acyl chain in formation can only contain up to 12–14 carbons, thus limiting the size of the de novo synthesized FA in the apicoplast20. De novo synthesis of myristic acid was shown to be essential for tachyzoite intracellular development, as it is used to generate LPA, then exported to make the bulk of parasite phospholipid content and allow parasite membrane biogenesis and propagation12,14,16,21. Furthermore, Toxoplasma’s genome contains a single cytosolic N-myristoyltransferase (NMT), which is responsible for protein myristoylation as a post-translational modification that affects their cellular localization, signaling events, and protein-protein interaction, for parasite survival22. In silico study predicted more than 150 myristoylated proteins22 and a recent study identified more than 42 NMT substrates23. Among them, the myristoylation of TgMIC7 was shown to be critical to parasite invasion and survival. Therefore, myristic acid made in the apicoplast could also participate in protein acylation and thus would also need to be exported from the apicoplast to be utilized for NMT.
Despite the apparent importance of trafficking metabolites to and from the apicoplast, only a couple of apicoplast transporters have been identified24,25,26,27. Importantly, although essential chloroplast lipid transporters have been identified and characterized in plants, such as ABC transporters for PA import called TGDs28,29,30, and fatty acid exporters (FAXs)31,32. No homolog could be found in Apicomplexa, nor in their closest photosynthetic homologs, the Chromerids Chromera velia and Vitrella brassicaformis, suggesting their potential loss during evolution. Taken together, this leads to the central question: What is (are) the transporter(s) responsible for exporting lipids and FAs from the apicoplast of apicomplexan parasites?
P-Type ATPases are ubiquitous membrane-bound and selective transporters that use the energy from ATP hydrolysis to transport substrates across the plasma or organellar membranes. They are divided into five sub-families based on their substrate specificities33. P1-, P2-, and P3-ATPases are known as cationic transporters, such as the P2-ATPase known as SERCA, which pumps calcium into the ER in most eukaryotes34. The last two families, P4 and P5, are present only in eukaryotic cells and have so-called “giant substrates.” P4-ATPases are known as amino-phospholipid flippases that can transport lipids across biological membranes with key roles for lipid trafficking, lipid homeostasis, and membrane asymmetry. Their presence and essential roles have recently been revealed in Toxoplasma, notably for microneme secretion and parasite invasion/egress35,36,37,38,39,40. The last and least characterized sub-family, the P5-ATPase, has no defined canonical substrate. The P5-ATPase is currently divided into two clades: the P5A and P5B, which localize to the ER membranes, and the vacuole or lysosome membrane, respectively33.
Here, we characterize a novel putative P5-ATPase transporter in Toxoplasma gondii, named TgFLP12, which localizes to the apicoplast and is important for parasite development. Lipidomic, lipid sources analyses by stable isotope labeling, substrate complementation, and heterologous complementation approaches revealed that, unlike plant and green algal chloroplasts, the apicoplast of T. gondii uses this P5C-ATPase TgFLP12 as its FA exporter. This further reveals the complex metabolic pathways put in place by Apicomplexa through their evolution as obligate intracellular parasites to provide essential lipids for their propagation.
Results
TgFLP12 is a P5-ATPase belonging to a new clade conserved in Apicomplexa but not in higher plants
We hypothesized that the apicoplast FA transporter might fall in the P-type ATPase transporter family, to which belong known phospholipid lipid “flippase” transporters. To identify such possible ATPase involved in the apicoplast lipid transfer in Toxoplasma gondii, we screened its genome using the sequence of known P-type ATPases from Saccharomyces. Several P-type transporters have already been identified and characterized in Toxoplasma, such as the ER-resident P2-ATPase transporter TgSERCA, located in the ER membrane41, the P3-ATPase PMA1 involved in bradyzoite differentiation42, as well as several lipid P4-ATPases involved in parasite microneme secretion, invasion, and egress39,40,43.
This approach allowed us to identify a total of 18 putative P-Type ATPases in T. gondii (Fig. 1, Supp. Figure 1): eight putative cation transporters belonging to families P1, two, and three types ATPases, six putative P4-ATPAses/lipid flippases, and two putative P5A/B ATPases. The P5A clade members have been localized into the ER membrane where they can export mistargeted mitochondrial44 or import small Wnt proteins45. The P5B clade ATPases, usually localize at vacuolar or lysosomal membranes, and recent data show that one member could transport polyamine-containing molecules (spermidine/spermine) from the lysosome to the cytosol46,47.
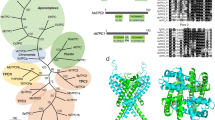
Maximum likelihood phylogeny of P5-ATPases. The tree was derived by using the best-fit model (LG + F + R7) in IQ-TREE and shows two sets of UFBoot2 supports at branches (1000 replicates). The first set corresponds to P5-ATPase only (114 sequences as shown here) and the second set corresponds to a global dataset of P-type ATPase (207 sequences; Fig. S1). The rooting of the P5 clade is shown by the dashed line. Black dots indicate 100/100 support. Dashes indicate support <80 or a different topology in one of the analyses (supports are not shown where <80 in both analyses).
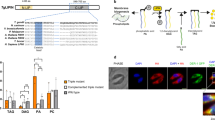
Interestingly, the phylogenetic screening identified two peculiar P-type ATPases of which, TGGT1_236860 and TGGT1_289070, the latter of which we named TgFLP12 due to the presence of twelve putative transmembrane domains. TgFLP12 bore all typical cytosolic domains found in P-Type ATPases, the actuator (A)-___domain (Fig. 2a and b, light blue), the nucleotide-binding ___domain (orange), and support ___domain (green) (N-___domain and P2-___domain, respectively) and the Phosphorylation ___domain (P1-___domain) (Fig. 2a and b).
a Cytosolic P-type ATPase conserved domains, A-___domain = Actuator ___domain (blue), N-___domain = Nucleotide ___domain (orange), and P1- and P2-domains = Phosphorylation ___domain (green and black). b Phyre2 3D model based on yeast P5-ATPase homology and obtained with ChimeraX 1.2.5, blue = A-___domain, orange = N-___domain, and green = phosphorylation site. cTgFLP12-HA apicoplast localization was determined by IFA with intracellular markers and observed by epifluorescence (top panel) or confocal (lower panel) microscopy. Blue = Hoechst, Red = apicoplast markers (Stromal CPN60 and outermost membrane ATRx1), and green = 3xHA tag. Observations were performed on at least three biological replicate experiments.
Surprisingly, phylogenetic analysis showed that TgFLP12 does not cluster with any of the previously mentioned P5A- or P5B-ATPases but rather to a third and newly identified clade that we called P5C-ATPase. The P5C clade is absent from animals, yeasts, and flowering plants, but is present in a range of other eukaryotes including fungi, basal land plants, such as mosses, and numerous protists, such as diatoms, oomycetes and alveolates (Fig. 1). We identified homolog in Plasmodium falciparum (PF3D7_0504000) and showed that both proteins belong to the P5C-ATPase subfamily. Interestingly, it seemed that the gene has independently duplicated in Cryptosporidium (an Apicomplexan parasite devoid of apicoplast) and coccidians, both bearing different copies of the gene with different predicted localization48. P5-ATPases are known to be subdivided into two clades. This data suggests that TgFLP12 is a novel P5C-ATPase in Toxoplasma gondii that could transport some type of macromolecules.
TgFLP12 is an apicoplast membrane protein
The localization of TgFLP12 was unclear based on existing information. TgFLP12 has no predicted N-terminal targeting sequence, such as the bipartite signal characteristic of apicoplast targeting. Its predicted localization by HyperLOPIT (Barylyuk et al.48, ToxoDB.org) suggests that it would be a Golgi and/or Plasma membrane protein whereas immunofluorescence assay (IFA) on its murine malaria agent (P. berghei) ortholog revealed an apicoplast localization25. Thus, to determine the actual localization of TgFLP12 in vivo in Toxoplasma, we generated an HA-tagged TgFLP12 parasite line by inserting the coding sequencing of a triple hemagglutinin (3xHA) epitope-tag to the 3’ end of TgFLP12 gene using the pLIC strategy49, validated by a PCR screen (Supp. Fig. 2a and b). IFAs revealed that TgFLP12-HA co-localized with the apicoplast stromal marker, chaperonin 60 (CPN60) (Fig. 2c) similar to the P. berghei homolog25. More specifically, an accumulation of the HA signal on the periphery of the apicoplast is observed (Fig. 2c), suggesting localization in the apicoplast membranes, and correlating with the presence of predicted transmembrane domains. To confirm this hypothesis, further IFAs were conducted with the apicoplast outermost membrane marker, Apicoplast Thioredoxin 1 (ATRx1) using confocal microscopy. This confirmed the apicoplast localization and revealed a robust colocalization of ATRx1 with the 3xHA signal, Pearson’s coefficient of 0.80 (Fig. 2c, lower panel), strongly pointing at an apicoplast membrane localization of TgFLP12. Moreover, TgFLP12 was recently pulldown after addition of the Turbo-ID element on the apicoplast outer-membrane protein, Apicoplast Phosphate Translocator50, supporting its localization to the apicoplast membranes. Western-blot analysis further confirmed the predicted molecular weight of TgFLP12 at ~220 KDa (Supp. Fig. 2e).
TgFLP12 is important for tachyzoite development and apicoplast integrity
To investigate the functional role of TgFLP12, and following its negative phenotypic score (−3.25)51,52, an inducible knock-down strain was generated. Using the promotor replacement TATi-TET system facilitated by a CRISPR-Cas9 approach35 (Supp. Fig. 2a) in the TgFLP12-HA strain we obtained the TgFLP12-HA-iKD strain. Integration of the exogenous promoter to the correct locus was confirmed by PCR (Supp. Fig. 2d and e). The depletion of the TgFLP12 protein was confirmed by western blot after 48 h of anhydrotetracycline (ATc) treatment (Fig. 3a).
a Inducible knockdown of TgFLP12 in the TgFLP12-HA line after promoter replacement. TgFLP12 was detected by Western Blot using Rt anti-HA antibody, and complete shutdown of the protein occurred after 48 h of ATc treatment. TOM40 (lower panel) served as loading control. Illustrative WB from at least three biological replicates. b Detrimental effect of TgFLP12 loss shown by plaque assays performed in the absence (-) or presence (+) of ATc and with different FBS concentrations (0, 1 and 10%) and fixed after 7 days. c Plaque size quantification and statistic from plaque assay (b). Data are presented as mean values +/− SD (n = 3). d The absence of TgFLP12 reduced the intracellular replication of Toxoplasma tachyzoites. Two-way Anova, Šídák’s multiple comparisons, data are presented as mean values +/− SD (n = 4.) e Apicoplast disruption in absence of TgFLP12 revealed by IFA and using an anti-CPN60 antibody, Blue = Hoechst, Red = apicoplast marker CPN60, and green = 3xHA tag. f Statistical analysis of the apicoplast disruption after complete shutdown of TgFLP12 expression. Data are presented as mean values +/− SD (n = 3). g Transmission electron microscopy showing apicoplast morphology after 48 h of ATc treatment on TgFLP12-iKD, apicoplast appeared swelling and accumulating electron light material in the stroma (redhead arrows) as well as in the intermembrane spaces (red arrows). Images are illustrative of average observations on at least three biological replicates.
We, and others, have recently revealed that the parasite can adapt its metabolic program to host nutrient content and that some proteins become more or less important if the host nutrient content is higher or lower6,7,16,19,53. To assess whether TgFLP12 had a more specific role in cases under fluctuating nutrient content, TgFLP12-iKD was grown under high, normal, or low nutrient/lipid content at 10, 1, or 0% FBS, respectively, as previously described6. We performed plaque assays and found that the disruption of TgFLP12 induced a drastic, and almost complete, reduction of the plaque size in the presence of ATc, demonstrating the importance of the protein for intracellular development (Fig. 3b and c). Interestingly, increase in nutrient content, with increasing concentrations of FBS slightly compensated for the phenotype of TgFLP12 disruption (Fig. 3b, c). As a negative control, the ATc treatment showed no effect on parasite development and plaque size in the parental strain (Supp. Fig. 2f).
To further assess the importance and role and TgFLP12 for parasite intracellular development, we performed replication assays. The protein down-regulation takes about 48 h to be complete, as controlled by Western Blot (Fig. 3A), so the mutant was pre-treated for 3 days with ATc for a complete loss of TgFLP12. Parasites were collected and allowed to infect new HFF for 24 h with ATc, to be compared to the WT. Tachyzoite replication was significantly impacted upon the disruption of TgFLP12: there was a significant decrease in the number of large vacuoles with a drop from ~65 to ~45% for vacuoles with 8-stage parasites and ~20 to ~6% for vacuoles with 16 parasites, and a significant accumulation in small vacuoles (1-, 2- and 4-stage parasites) in the absence of TgFLP12 compared to with TgFLP12 present (Fig. 3d). These data demonstrate that TgFLP12 depletion reduces Toxoplasma division rate after complete loss of the protein and therefore reduces parasite fitness in the long term.
Since TgFLP12 is essential for parasite growth and localizes to the apicoplast, we sought to determine whether the depletion of TgFLP12 could impact the biogenesis, morphology, or presence of the organelle. To do so, similarly as for the replication assay, parasites were pre-treated for 72 h with ATc and allowed to re-invade fresh HFF for an extra 24 h. We then conducted IFA using the stromal apicoplast marker CPN60 to scrutinize the apicoplast (Fig. 3e). In WT parasites (i.e., TgFLP12-HA-iKD in the absence of ATc), the apicoplast forms a single dot per parasite located on the apical part of the parasite. The addition of ATc, which suppresses TgFLP12, led to the loss of the apicoplast in most parasites within the same vacuole and/or a swelling of the few remaining (Fig. 3e, lower panel red head arrow). Furthermore, the apicoplast was also sometimes found at the residual body (Fig. 3d, lower panel red arrows). This mislocalization of CPN60 in the TgFLP12-depleted parasites was quantified, and 60% of vacuoles presented a CPN60 signal that localized to the residual body (Fig. 3f). Together, this data demonstrates that TgFLP12 is essential for Toxoplasma tachyzoite development and the maintenance of the apicoplast. Whilst the apicoplast is disrupted by TgFLP12 deletion, Toxoplasma mitochondria were not affected after ATc treatment and TgFLP12 loss, as the intracellular parasites have conserved their lasso shape-like mitochondrial form (Supp. Figure 3). Similar results were previously obtained in the apicoplast ACP (a key enzyme of the FASII pathway) mutant: the deletion of the ACP protein led to the disruption of the apicoplast but the mitochondria remained unchanged2.
Further analysis by transmission electron microscopy on parasites treated and untreated with ATc for 48 h, revealed gross anomalies in the apicoplast morphology. Parasites depleted for TgFLP12 were mostly similar to WT parasites with normal shaped apical machinery (micronemes, rhoptries, and conoid), and no visible change in the mitochondria, ER, nucleus, or Golgi apparatus. However, the only change that could be observed was the drastic effect on the apicoplast: the organelle was found massively enlarged and often found with the accumulation of low-density material within the stroma (redhead arrows) as well as between the membranes (red arrows; Fig. 3g). Down-regulation of TgFLP12 rapidly led to a morphological change of the apicoplast within 48 h (Fig. 3g) that resulted in the longer-term, to the loss of the apicoplast (Fig. 3e, lower panel). Together, this suggested that the disruption of TgFLP12 has a major and central effect on the apicoplast maintenance, and possibly the accumulation of material within.
TgFLP12 is a predicted macromolecule transporter, but its substrate and function remain a mystery. Given TgFLP12 localizes to the apicoplast, the site of de novo lipid synthesis, we decided to determine the impact of TgFLP12 loss on the parasite lipid content.
TgFLP12 depletion alters parasite lipid homeostasis and FA metabolism
To determine whether TgFLP12 could impact the parasite lipid content and homeostasis, we performed mass spectrometry-based lipidomic analysis of our TgFLP12-HA-KD mutant. TgFLP12-HA-iKD are cultured during 72 h with ATc ( + ATc, green bars), harvested and compared to the untreated (-ATc, grey bars) (Fig. 4a). The total FA abundance (comprising all FA from membrane/storage lipids as well as free FA (FFA)) did not significantly change in absence of TgFLP12 (Fig. 4a) despite a small increase in the total FA in TgFLP12-KD. However, the detailed FA composition (i.e., the different molecular species of FA found in the parasite where FA are commonly labeled Cx:y, x being the number of carbons that made the chain and y the number of unsaturated bonds) revealed important significant changes in the FA homeostasis of the parasite in the absence of TgFLP12.
a FA total amount, quantified by GC-MS is unchanged after iΔTgFLP12 repression. Data are presented as mean values +/− SD (n = 4). b FA composition in Mol% of TgFLP12 lacking parasites ( + ATc) compared to parasites harboring TgFLP12. Two-way Anova, Šídák’s multiple comparisons, data are presented as mean values +/− SD (n = 4). c Lipid composition revealed by one-dimensional TLC and GC-MS showed a decrease in phospholipids and an increase in neutral lipid TAG and DAG in parasites without TgFLP12 (+ATc) compared to parasites with TgFLP12, one sample t test, data are presented as mean values +/− SD (n = 4). Major FA relative abundance of d triacylglycerols TAG, e diacylglycerols DAG, f phospholipids PL, and g free fatty acids FFA. Two-way Anova, Šídák’s multiple comparisons, data are presented as mean values +/− SD (n = 4). h Relative abundance of the different lipid classes made from scavenged lipids revealed that neutral lipids TAG and FFA from scavenged lipids increase after addition of ATc. One sample t test, data are presented as mean values +/− SD (n = 4).
Measure of the relative abundance of each individual FA (mol%) revealed a significant decrease of C14:0, myristic acid (Fig. 4b), i.e., the major FA product of the apicoplast FASII14,54, which is usually reduced following the loss of the apicoplast and its FASII. However, there was an unusual and significant increase in the content of C16:0, palmitic acid (Fig. 4b), i.e., the second major product of the apicoplast FASII. This is a very unique and surprising increase, which has never been observed and reported in previous disruptions of apicoplast proteins that usually reduce the presence of C16:0, along with C14:014,19,50,54,55,56,57. This unusual accumulation of C16:0 was further confirmed by conducting 13C-U-acetate labelling to monitor parasite FA elongation58, which showed that C16:0 was significantly more used as a FA substrate to elongate parasite FA, in the absence of TgFLP12 (Supp. Fig. 4a). As mentioned earlier, these changes were solely measured in the absence of TgFLP12, unlike any previous apicoplast protein analyzed thus far14,19,50,54,55,56,57 and cannot be attributed to the addition of ATc as there was no change in the parental strain FA composition under ATc treatment (Supp. Fig. 4b and c).
To further understand the impact of the loss of TgFLP12 on parasite lipid content, major lipid classes were separated using high-performance thin layer chromatography (HPTLC), i.e., PL, the most abundant membrane lipids, TAG, and diacylglycerol (DAG) together making the bulk of storage lipids, and FFA, the hydrophobic building blocks of most lipid classes. Quantification and comparison of each of these classes using GC-MS analyses, revealed a strong significant increase of ~2.5-fold in TAG content, coupled with a milder yet significant increase of DAG, together pointing at an increase of lipid storage (Fig. 4c). On the other hand, a mild but significant decrease in PL content was observed, whilst FFA remained unchanged (Fig. 4c). Separating the different phospholipid species revealed that This decrease in PL content appeared to correspond to a significant decrease in phosphatidylcholine (PC, i.e., the most abundant membrane phospholipid) relative abundance, while no other PL class was affected (Supp. Fig. 4d).
Furthermore, detailed analyses of the molecular species for each major lipid class by GC-MS revealed a common change for most parasite lipid classes (TAG, DAG, FFA, PL): a significant decrease of C14:0, typically made by the apicoplast FASII, in the absence of TgFLP12 (Fig. 4d–g), further supporting our hypothesis of the lack of transport of this apicoplast FA product.
The strong increase in storage lipids in the absence of TgFLP12 potentially indicated that the parasite could be scavenging more lipids from the host, then channeled to the parasite lipid storage, as we previously showed6. To test this hypothesis, host-scavenged FA flux toward the parasite was measured by stable isotope labeling combined with GC-MS analysis, as previously reported6. Briefly, host cells were grown with media containing 13C-U-Glucose and grew until confluence, in the absence of parasites. Host FA can thus be naturally synthesized fully labeled with 13carbons via the action of the host cytosolic FASI. The media is then replaced with normal (unlabeled glucose), parasites are added and allowed to grow on pre-labelled host cells. After 72 h of infection, parasites are separated from host cells debris by syringing, then filtering, and parasite FAs are analyzed by GC-MS. Any FA labeled with 13carbon are assumed to be scavenged from the host (Supp. Fig. 5a), as previously controlled and confirmed6,59,60.
Measuring parasite scavenging confirmed that most FAs from parasite TAG in the absence of TgFLP12 were directly scavenged by the parasite from the host cell, whereas it was not the case for DAG and PL (Fig. 4h). More surprisingly, while the level of FFA remained the same in absence of TgFLP12, the FFA sourced from the host increased by 5-fold (Fig. 4h).
This showed that loss of TgFLP12 impacts the parasite lipid homeostasis. Despite the unchanged absolute level of C14:0 in increased C16:0 (Fig. S4), relative composition shows that there is a significant global decrease in C14:0 content (Fig. 4b, d-g). While this suggests that the apicoplast FASII is still active, we measured that the parasite further increases lipid scavenging (Fig. 4g), which is usually happening when the FASII products are lacking16,17,19, further pointing at an issue with the availability of apicoplast FA. All point to a potential deficiency at the level of the apicoplast-derived FA, and its capacity to provide FA to the parasite.
Disruption of TgFLP12 impacts the exit of myristic acid from the apicoplast
To test whether TgFLP12 is linked to de novo FA synthesis via the apicoplast FASII, we performed the stable isotope labeling using 13C-U-Glucose combined to GCMS-based lipidomic analyses to measure and quantify FASII activity in intracellular parasites6,54,61. Briefly, unlabeled confluent cells (i.e., that no longer perform de novo FA synthesis) are infected by parasites in a culture media containing 13Carbon-U-Glucose. The parasite scavenges this glucose and uses it to fuel its FASII activity, being the only pathway generating 13C-labeled FA, as previously reported6,14,19,58,61. After 72 h, parasites were isolated, and FA content analyzed by GC-MS (Supp. Fig. 5a). Results showed similar incorporation of labeled 13C into de novo synthesized FA in both WT and TgFLP12 mutants indicating that the apicoplast FASII pathway, per se, is still active in the TgFLP12 mutant during the 72 h of incubation with 13C-U-Glucose (Fig. 5a). More surprisingly though, The abundance and synthesis of the FASII-signature FA C14:0 were unchanged, and in some FA, such as C16:0, and C18:0 (second and third products of the FASII in abundance14,54), 16:1 and 18:1 (direct down products of the FASII via the ER elongation pathway58), the abundance of 13C labeled FA was higher in the mutant, suggesting there is some alteration of FASII activity or the fate of resulted FAs from the FASII (Fig. 5a).
a %13Carbon incorporated by parasite de novo FASII in TgFLP12-iKD FA in presence and absence of ATc after 72 h incubation 8 mM 13C-U-Glucose. Two-way Anova, Šídák’s multiple comparisons, data are presented as mean values +/− SD (n = 4). Mass isotopomer distribution of b C14:0 and c C16:0 showing the incorporation of 13C by FASII 72 h into the FA chains, M+ indicates the number of 13carbon incorporated per chain. Two-way Anova, Šídák’s multiple comparisons, data are presented as mean values +/− SD (n = 4). d %13Carbon incorporated by parasite de novo FASII in TgFLP12-iKD FA in presence and absence of ATc of ATc after 24 h incubation with 8 mM 13C-U-Glucose after complete loss of TgFLP12 = 48 h incubation with ATc prior to 13C-U-Glucose addition. Two-way Anova, Šídák’s multiple comparisons, data are presented as mean values +/− SD (n = 7). Mass isotopomer distribution of e C14:0 and f C16:0 showing the incorporation of 13C by FASII 24 h into the FA chains, M+ indicates the number of 13carbon incorporated per chain. Two-way Anova, Šídák’s multiple comparisons, data are presented as mean values +/− SD (n = 7). g %13Carbon incorporated by parasite elongation in TgFLP12-iKD FA in presence and absence of ATc after 24 h incubation with 8 mM 13C-U-Acetate after complete loss of TgFLP12 = 48 h incubation with ATc prior to 13C-U-Glucose addition. Two-way Anova, Šídák’s multiple comparisons, data are presented as mean values +/− SD (n = 4). Mass isotopomer distribution of h C16:0 and i C18:0 showing the incorporation of 13C by elongation 24 h into the FA chains, M+ indicates the number of 13carbon incorporated per chain. Two-way Anova, Šídák’s multiple comparisons, data are presented as mean values +/− SD (n = 4). jTgFLP12-iKD FA abundances with 13Carbon incorporated by the host and scavenged by the parasite in presence and absence of ATc. Two-way Anova, Šídák’s multiple comparisons, data are presented as mean values +/− SD (n = 4).
Although the abundance of C14:0 made by FASII during 72 h did not alter, its mass isotopologues distribution (MID) revealed major changes from the WT profile (Fig. 5B). Both WT and mutant (72 h incubation with 13C-U-Glucose) had the 13C incorporation up to the M + 14, fully labeled with 13C, suggesting the FASII is fully active in both WT and TgFLP12 mutant, with a two-by-two increase of the mass, typical of the FASII activity, showing the full synthesis of myristic acid, C14:0. However, in the mutant, the fully labeled FA isotopomer species, i.e., C14:0 M + 14, is significantly reduced compared to that of WT. This is consistent with the lower abundance of C14:0 previously measured (Fig. 4b). Similarly, in C16:0 MID, as well as most other FA species, the mutant contained a higher abundance of the M + 14 species, and M + 16 than that of WT, as if M + 14 isotopomer accumulated in the absence of TgFLP12 (Fig. 5c, Supp. Fig. 6a–d). Interestingly, this phenotype is, again, unique to the disruption of TgFLP12, unlike disruption of the FASII pathway and/or the apicoplast has always led, thus far, to the significant decrease of all apicoplast FA products/isotopologues, especially C14:0 and C16:0, and their derived elongated products14,19,50,54,55,56,57.
This suggests two possibilities: (1) Myristic acid, C14:0, underwent another round of elongation through the FASII pathway instead of being exported as such, and then was exported outside of the apicoplast once elongated to a C16:0 palmitic acid-containing product (e.g., LPA C16:0). (2) The increase of FASII activity could have occurred during the 72 h of TgFLP12 downregulation. Indeed, since 48 h of ATc treatment are required to shutdown TgFLP12 expression and to get rid of the remanent proteins, slow disruption of TgFLP12 coupled with its impact on parasite lipid homeostasis, could have triggered an initial increase of FASII activity, as previously reported14, and its arrest once apicoplast was impacted after 48 h.
To test this second hypothesis, parasites were pre-treated with ATc for 48 h to completely deplete TgFLP12 before 24 h incubation with 13C-U-Glucose, parasites harvest, and lipid analysis (Supp. Fig. 5b). This first showed that there was incorporation of 13C, proving that the FASII is still active after full disruption of TgFLP12. Interestingly, the incorporation of 13C showed a general decrease for most FA species and most significantly for C14:0, C16:0, C16:1, and C20:1 (Fig. 5d). This confirmed that FASII activity increases during the loss of TgFLP12 as previously observed for 72 h (Fig. 5a). However, full depletion of the protein before 13C incubation induced a decrease in the FASII activity. Accordingly, MID profiles of C14:0 and C16:0 (Fig. 5e and f) revealed a significant decrease in the 13C incorporation for all isotopologues. This was further confirmed for most FA typically made from FASII (Supp. Figure 6e, f, g, and h).
Previous studies showed that FASII products, C14:0/C16:0, exit the apicoplast to reach the ER to be further elongated14,54,58. Furthermore, our data points to an increase in de novo synthesis of C16:0 in the apicoplast and putatively from the ER elongation pathway. To test this further hypothesis and to determine whether the disruption of TgFLP12 had an impact on ER FA elongation, parasite FA elongation activity was measured by culture with 13C-U-Acetate, (which is used by the parasite ER elongases (ELO A, B, and C) to elongate FA by two-carbon units58), followed by GC-MS analysis (Supp. Figure 5b). As for the measurement of FASII activity, the elongation had been measured for 24 h after complete repression of TgFLP12, to avoid acetate degradation. After 24 h of incubation of 13C-U-Acetate, parasite elongation was still active in total absence of TgFLP12. However, fewer 13Carbons were incorporated in C16:0, C16:1, and C20:1, while more C18:0 has been elongated from the elongation pathway (Fig. 5g). Interestingly, MID of C16:0 FA revealed a significant decrease of C16:0 made from shorter chains, mainly from the typical apicoplast C14:0, with a significant decrease in M + 2 in the mutant (Fig. 5h). This corroborates the previous hypothesis that C14:0 cannot exit the apicoplast to be elongated in the ER in the TgFLP12 mutant. In parallel, MID of C18:0 FA (Fig. 5i) revealed that more have been elongated directly from C16:0 chains, instead of the usual C14:0. This was also true for the C18:1, C20:0, C20:1, C22:0, and C24:1. Taken together, this suggest that most elongated FAs seem more generated from a C16:0 precursor than from C14:0, again pointing at the reduced C14:0 content in the ER, site of FA elongation (Supp. Fig. 7c, e, f, g, and h respectively).
As previously reported, Toxoplasma relies on both FA scavenging and de novo FASII synthesis to generate FA, the activity of each pathway can be up- or down-regulated based on host nutrient and FA availability as a balance between the two6,19. In this study, TgFLP12 disruption induces a boost in FASII activity prior to a reduction (Fig. 5a, d), a lack in C14:0, an increase in C16:0 and more storage lipid originated form the host (Fig. 4h). To determine whether the parasite switched FA acquisition from FASII de novo synthesis to host scavenging, we monitored host scavenging by metabolic labeling and GC-MS analyses. This revealed that the disruption of TgFLP12 for 72 h induces a significant increase of C18:1 scavenged from the host. We also observed a trend of increased scavenging for most major FA species C14:0, C16:0, C16:1, and C20:1 form the host (Fig. 5j).
The disruption of TgFLP12 results in a specific loss of FASII-made C14:0 FA, and unusual de novo synthesis of C16:0 product, and an increase usage of that C16:0 for FA elongation. TgFLP12 depletion does not behave like any major protein that disrupts the apicoplast and its FASII14,19,50,54,55,56,57: its depletion does not directly arrest FASII activity, which rather increases during the first moments of the protein loss, then slows down after the disruption of the apicoplast, concomitantly with an increase of host FA scavenging for TAG storage. Together with the putative role of exporter, lipidomic and lipid flux analyses suggest that TgFLP12 is linked to the activity of apicoplast FASII and/or the trafficking of its products from apicoplast to the ER.
TgFLP12 is responsible for FFA export from the apicoplast
TgFLP12 deletion impacts the apicoplast FA de novo synthesis, ER elongation, and scavenging the host, with as global significant decrease of C14:0. Thus, to further delineate the role of TgFLP12 for lipid derived from FASII, the amount of FASII-derived FA in major lipid classes were quantified using 13C-U-Glucose labeling, 1DTLC, and GC-MS analyses. No difference was observed in the global abundance of PL, TAG, and DAG originating from FASII. However, the abundance of FFA sourced by the apicoplast FASII was significantly reduced by 30% in the TgFLP12 mutant (Fig. 6a). Moreover, the incorporation of 13Carbon by FASII was significantly reduced in the mutant only for C14:0 of FFA (Fig. 6b). The same phenotype was observed for DAG and PL made from FASII, where C14:0 was significantly reduced but not for TAG, which depends more on scavenging, especially in the absence of TgFLP12 (Supp. fig. 8a–c). These different elements all suggested that TgFLP12 facilitates apicoplast FASII product (C14:0) to exit the apicoplast and provide the bulk of FFA to fuel lipid storage and membrane biogenesis.
a Relative abundance of TAG, DAG, PL, and FFA originated from parasite FASII over 72 h. One sample t test, data are presented as mean values +/− SD (n = 4). b Percentage of 13C incorporation in major FA species of FFA made by parasite FASII over 72 h in presence (-ATc) and absence ( + ATc) of TgFLP12. Two-way Anova, Šídák’s multiple comparisons, data are presented as mean values +/− SD (n = 4). c Plaque assays complemented with possible TgFLP12 substrates, FFA C14:0 or C16:0, and LPA C14:0 or C16:0. d Plaque size quantification and statistics from plaque assay (c). Two-way Anova, Tukey’s multiple comparisons, data are presented as mean values +/− SD (n = 3). e and f FA composition in Mol% of yeast WT + Ф (empty vector), fatΔ+Ф (empty vector) and fatΔ complemented with vector containing TgFlp12 gene cultured with D31-C16:0 (e) or D33-C18:1 (f). Two-way Anova, Tukey’s multiple comparisons, data are presented as mean values +/− SD (n = 3).
The apicoplast is the place for essential metabolic pathways, and to date, none of the exporters putatively responsible for the exit of the down-products of these pathways have been identified27. Previous studies showed that most de novo apicoplast pathways could be rescued by exogenous metabolite complementation. FASII can be rescued artificially by exogenous addition of C14:0, and/or C16:016,17,54, LPA synthesis by exogenous addition of LPA C14:014, and the heme pathway by 5-ALA16. Moreover, recent studies on P5B-ATPases have shown some to be polyamine (spermine/spermidine) transporters46,47. Using the same media complementation strategy, we decided to perform plaque assays supplemented with known scavenged metabolites, and if we can rescue the growth in ΔTgFLP12, we should identify the product transported by TgFLP12.
Polyamines are essential for Toxoplasma survival and can be scavenged from the host62. Experiments showed that spermidine and spermine did not complement TgFLP12 function, nor did the heme pathway intermediates 5-ALA and PPIX (Supp. fig. 8d), indicating TgFLP12’s lack of involvement in polyamine and heme trafficking.
This study suggests TgFLP12 facilitates the export of lipid products from the apicoplast, particularly lipids with C14:0 FA chains. Exogenous LPA (C14:0) and LPA (C16:0) slightly improved parasite growth (Fig. 6c, d). Some plaques were observed but did not specifically rescue the absence of TgFLP12, whereas FFA C14:0 and C16:0 significantly rescued TgFLP12-deficient parasites with no positive effect on WT parasite growth, unlike LPA complementation. This strongly suggests that the FFA rescue effect is directly due to the complementation of TgFLP12, and thus likely indicates that TgFLP12 exports FA products from the apicoplast FASII pathway. The rescue effect was stronger for FFA C14:0 than C16:0, highlighting TgFLP12 specificity.
Finally, to further validate and confirm the FA exporter activity of TgFLP12, we performed an established heterologous complementation that was used to identify the plant chloroplast FA exporter FAX131, combined to an import assay of deuterated FA, which revealed the function of both yeast and murine FA transporters63,64,65. Briefly, we used the yeast strain lacking Δfat1 one of the yeast plasma membrane fatty acid transporters. The Δfat1 was either complemented with a copy of TgFLP12 or an empty vector (Supp. fig. 9). We then measured the import of exogenous deuterated (D-)C16:0 (D31-C16:0) or deuterated C18:1 (D33-C18:1) into the yeast cells pellets from the WT strain, the fat1Δ mutant, and the TgFLP12 complemented strain, fat1Δ+TgFlp12. The yeast fat1Δ has a significant decrease in the import of D-C16:0 compared to WT, as previously reported63. However, the Δ fat1Δ import capacity was restored to a higher level than WT when complemented with TgFLP12 (Fig. 6e), confirming the FA transport capacity of TgFLP12. We noticed that the abundance of C16:0 was increased in the fat1Δ strain compared to WT, that was certainly due to an increase FAS activity due to reduce import of exogenous C16:0. Its abundance is also restored to the WT level in our complemented strain. Interestingly, in contrast, while the fat1Δ strain had no reduction of import of D-C18:1, complementation with TgFLP12 did not increase the uptake of this deuterated lipid (Fig. 6f).
Taken together, the depletion of TgFLP12 induces a loss of the apicoplast, similar to other known apicoplast mutants, however the resulting defect caused by loss of TgFLP12 in its lipidome is unique amongst other apicoplast mutants14,19,50,54,55,56,57. TgFLP12 disruption induces a significant reduction in intracellular development, which is partially compensated by a higher host nutrient environment, correlating with previous results on the balance between de novo FA synthesis and host scavenging14,16. The loss of TgFLP12 specifically induces a severe reduction of the signature FASII FA C14:0 in most parasite lipid species, except for TAG as scavenging capacities increase in the absence of TgFLP12 (Fig. 7). It further induces a specific accumulation of C16:0 that is extensively used to elongate FASII products. These complementation assays consolidate the lipidomic data, which indicate that TgFLP12 plays a specific role in the apicoplast by exporting lipids originating from the FASII pathway outside of the apicoplast, with a higher affinity for FFA than LPA and a higher affinity for C14:0 length chain compared to C16:0 chain. Finally, exogenous complementation and heterologous complementation validated TgFLP12 as the exporter of apicoplast FA products from the FASII, most likely myristic acid, C14:0, which are then exported toward the ER for further elongation.
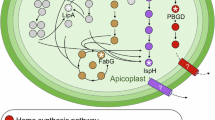
FASII pathway generates short FA chains, C14:0. From Amiar et al. 14, we know that apicoplast FAs can be esterified on a G3P backbone by ATS1 to form LPA, which exit the apicoplast to reach the ER via an unknown pathway. This exported LPA serves for phospholipids synthesis and FAs are elongated by the three ER elongases ELO A/B/C. This study suggests the role of TgFLP12 as an FFA exporter that allows the exit of FA C14:0/C16:0 from the apicoplast to ER to be elongated/desaturated and incorporate in bulk PL or storage lipids preventing toxic accumulation and disruption of the apicoplast. G3P = glycerol-3-phosphate; FASII = fatty acid synthesis pathway type II; FFA = free fatty acid; ATS1 = acyltransferase 1; LPA = lysophosphatidic acid; ELO = elongases.
Discussion
Despite the essentiality of the apicoplast-resident metabolic pathways, little is known concerning the different metabolite importers and exporters involved in these pathways27. Apicomplexa and their photosynthetic relatives Chromerida have lost canonical chloroplast lipid transporters, such as FAX131 and FAX2 or the ABC transporter, TGD28,29,30,66. Here, our data showed that its disruption is deleterious for apicoplast integrity as well as overall parasite survival, which is compatible with other known mutants of the apicoplast involved in lipid synthesis/trafficking14,54. We comprehensively investigated the lipidome of parasites lacking TgFLP12 and showed its importance for short fatty acid chain myristic acid, C14:0, exit of the apicoplast. These results showed that TgFLP12 substitutes for FAX proteins present in the chloroplast and preferentially exports C14:0 FA outside of the apicoplast.
Deep phylogenetic analyses reveal thatTgFLP12 belongs to a newly identified clade from the sub-family of P5-ATPase never described before, which we called P5C-ATPases. Although absent in most laboratory model species, the new P5C-ATPase family is present in a broad range of protists and is strongly resolved in the phylogeny, pointing to a potentially conserved substrate specificity different from the other P5 clades. The relationship between P5B-ATPases and P5C-ATPases remains unclear because P5B-ATPases do not form a strongly supported clade, and members of neither group have been functionally characterized in different taxa. The P5C-ATPase family shows several examples of secondary gene duplication events, including two that are specific to Cryptosporidium. The origin of the coccidian P5C-ATPase duplication is more difficult to pinpoint because deep relationships between apicomplexan sequences are not confidently resolved (support >95). While TgFLP12 is localized at the outermost membrane of the apicoplast, a LOPIT experiment in Toxoplasma predicted its paralog to localize to the rhoptries. Rhoptries are present in all apicomplexans, which may help to explain why P5C-ATPase is present in the apicoplast-lacking Cryptosporidium. Indeed, since P5C-ATPase is found in many non-photosynthetic eukaryotes (Fig. 1), the apicoplast FA transport in apicomplexans is apparently a derived state. Notably, P5A has been recently shown to transport proteins across the ER membrane45 and P5B to export polyamines from the lysosome to cytosol46,47. Given that the ER and lysosomal membranes are topologically equivalent with both, the membrane of (Golgi-derived) rhoptries and the outermost membrane of the apicoplast, it appears likely that P5 and P5C-ATPases were both ancestrally functionally connected with the endomembrane system. The presence FLP12 orthologs in all Alveolata with a functional plastid FASII strongly supports the role of FA exporter identified in Toxoplasma gondii, suggesting that this function might be conserved among other members of Alveolata, including photosynthetic red algae.
However, conservation of the FLP12 homolog in Apicomplexa lacking functional FASII raises the question of the FLP12 function in such organisms. Babesia and Theileria are two Piroplasmida, closely related to Haemosporidia Plasmodium spp, and for which FASII elements have not been identified67,68. These organisms also possess a life cycle within the blood as Plasmodium, in which FASII was shown to be dispensable during this stage69. A similar phenotype seems to happen in Babesia and Theileria, being in such a lipid-rich environment as the blood makes the apicoplast FASII dispensable. Moreover, these organisms have an important genetic diversity, and a genomes size reduced compared to Plasmodium (23 megabase vs Theileria: ~8 megabase; Babesia: ~14 megabase). It is possible that these organisms, with no need for FASII in blood stage, do not express (or very low) FASII elements during this stage, which could be the case in their final arthropod hosts (ticks) similar to Plasmodium that requires FASII in mosquito stage15. If FASII is indeed completely absent from Piroplasmida, FLP12 homolog might not have evolved to become an FA exporter, as mentioned previously, FA acid export seems to be a derived state of this new P5C clade as it is conserved in organisms without plastid.
This study reveals the essential role of short FFA originating from FASII to be exported outside of the apicoplast. We propose that TgFLP12 plays the role of the missing fatty acid exporter in the apicoplast, these exported FA can then be used by cytosolic/ER acyl-transferases19,70, such as GPAT for LPA synthesis, AGPAT for PA synthesis or DGAT for TAG synthesis and plays a role in forming the lipidic patchwork previously found during analysis of TgATS114. Our data from parasite elongation also revealed that in the TgFLP12 mutant, more elongation products were originating from a unique accumulation of C16:0 occurring in the absence of TgFLP12. Linked to previous data obtained for the TgATS1 mutant, we suggest that accumulated FFA C14:0 into the apicoplast undergo another cycle of elongation by the FASII and are then used by TgATS1 to produce LPA. Our elongation measurements support that FFA and LPA generated by the FASII can exit the apicoplast and then reach the ER where FA chains are elongated by ELO A/B/C (Fig. 6). However, this is still unknown how FFAs reach the outermost membrane, but several possibilities exist: (i) TgFLP12 localizes to the outermost membrane, but it is possible that TgFLP12 is inserted into the different membranes of the apicoplast. (ii) FFAs can also spontaneously insert into the apicoplast membranes and, thanks to the proximity of these membranes, naturally reach the outermost membrane where TgFLP12 export them to the ER. The apicoplast is known to also generate LPA and PA, but it remains unclear how these lipids exit the apicoplast.
The apicoplast has conserved essential metabolic pathways derived from a prokaryotic origin and has been shown to be an ideal drug target. However, whilst synthesis of essential metabolites has been well studied since the discovery of the apicoplast, very little is known about the transportation and trafficking of these metabolites. From TgFLP12 role in FA export and its importance for the apicoplast integrity, the production/export of LPA/PA could be a rate-limiting process in the apicoplast and TgFLP12 would have to act as a safety valve to prevent toxic FFA accumulation. Moreover, the importance of the apicoplast for the chronic persistence of parasite and bradyzoite viability was recently demonstrated71. This phenomenon emphasized the importance of investigating parasite metabolite transporters, which could certainly lead to the identification of promising therapeutic targets.
Methods
Toxoplasma gondii strains and cultures
HFF are cultured in Dulbecco’s Modified Eagle’s Medium (High Glucose DMEM, Life Technologies) supplemented with 10% FBS (Gibco), 2 mM glutamine (Gibco), supplemented with Pen/Strep and Fungizone at 37 °C and 5% CO2.
T. gondii tachyzoites (RH-Δku80, HA strains, and inducible knock-down strains) were maintained in HFF using Dulbecco’s Modified Eagle’s Medium (High Glucose DMEM, Life Technologies) supplemented with 1% FBS (Life Technologies), 2 mM glutamine and 25 μg/mL gentamicin at 37 °C and 5% CO2.
Generation of inducible knockdown FLP12 line in T. gondii
To introduce the endogenous tag at the 3’ end of the genes of interest, the open reading frames (ORF) of TgFLP12 were amplified by PCR. Amplicons were cloned into the plasmid LIC-3HA-CAT vector. After those plasmids were linearized with XbaI and transfected into the RH-Δku80 strain. An inducible knock-down strain was generated TgFLP12 by amplification of a mycophenolic acid resistance cassette, TATi sequence, and the inducible promoter TET from the plasmid pT8TATi1-HX-tetO7S1. For tag insertion or iKD, 50 μg of PCR products were co-transfected with 50 μg plasmid Sibley Cas9 coding for the endonuclease CRISPR-Cas9 and the appropriate protospacer (guide RNA) generated on the website platform CHOP-CHOP (chopchop.cbu.uib.no). To generate the TgFLP12-iKD described here, RH-Δku80 strains were first transfected with pLIC-TgFLP12-3HA, and then the promoter replacements for inducible knockouts were then transfected into cells with their respective 3’ HA tags. Electroporations were performed in a 2 mm cuvette in a BTX ECM 630 (Harvard Apparatus, at 1100 V, 25 Ω, and 25 μF). Stable lines expressing the constructs (endogenous tagging or exogenous promoter) were selected in media with 30 μg/ml chloramphenicol or 25 μg/ml mycophenolic acid and 50 μg/ml xanthine according to the resistance cassette of the insert transfected and cloned by limiting dilution.
Screenings of the parasite clones with correct HA element insertion and promoter replacement were done using primers pair in Table 1, supp Fig. 2. PCRs were performed with TaKara primestar max polymerase.
Immunofluorescence assay
Primary rabbit anti-CPN60 antibodies were used at a dilution of 1:500, rabbit anti-IMC1 at 1:500, rat anti-HA (Roche) at 1:500, and mouse anti-ATRx1 at 1:500. Secondary AlexaFluor 488- and 546-conjugated goat anti-rat and anti-rabbit antibodies (Life Technologies) were used at 1:1000 dilutions, respectively. Parasites were fixed in PBS containing 2% paraformaldehyde for 15 min at room temperature. Samples were permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature before blocking in PBS containing 2% FBS and incubation with primary antibodies then secondary antibodies diluted in the blocking solution. Labeled parasites were stained with Hoechst (1:10,000, Life Technologies) for 20 min and then washed three times in PBS then H2O. Coverslips were mounted onto slides before observation using a Zeiss epifluorescent microscope.
Phylogenetic analysis of P5-ATPases
We first used the TgFLP12 sequence to retrieve related entries from Uniprot by BLASTP and complemented this set by characterized P-type ATPases from the literature72,73,74. The preliminary dataset was expanded by additional BLASTP searches against GenBank nr and VEuPathDb. The sampling of the global P-type ATPase dataset was optimized to 207 sequences (Fig. S1) by several rounds of tree reconstruction in FastTree v2.1.11 (-lg -gamma parameters). A subset of 114 sequences comprising only P5-ATPases was then created from the global dataset (Fig. 1). All species present in this subset (except Thermochaetoides thermophila whose P5B sequence was added later from PDB) had their genomes exhaustively searched for P5-ATPase sequences. Final matrices for the global P-ATPase and P5-ATPase datasets were prepared by using the local pair alignment (-linsi) in MAFFT v7.505 and hypervariable site removal in BMGE v1.12 under relaxed criteria (-m BLOSUM30 -b 3 -g 0.4). Best maximum likelihood trees were computed by using the best fits models (LG + F + R8 for P-ATPase and LG + F + R7 for P5-ATPase) in IQ-TREE v2.2.0, each with 1000 UFBoot2 supports.
Western blot analysis
Proteins were harvested from scraped, needled (G26 needle), and filtered parasite cultures before counting by hemocytometer. Proteins were normalized and extracted by the addition of sample buffer to the parasite and boiled at 95 °C for 5 min. Equal volumes of protein samples were loaded and separated on a 4–12% gradient SDS-polyacrylamide (Life Technologies) and transferred to a nitrocellulose membrane. Incubation of the membrane with primary antibodies rat anti-HA (Roche) and rabbit anti-TOM40 followed by incubation with Horse radish peroxidase (HRP) goat anti-rat and anti-rabbit conjugated antibodies (Thermo Scientific). Revelation was done using the Biorad Chemidoc imager after incubation membrane staining with Luminata Crescendo Western HRP detection kit (Millipore).
Plaque assays in T. gondii mutants
Extracellular parasites were harvested after filtration and counted by hemocytometer. HFF monolayers were infected with ~1000 mutant parasites and allowed to develop under normal culture conditions or culture medium supplemented with 0.5 μg/ml ATc for 10 days untouched before staining with Crystal Violet (Sigma) and cell growth assessment by light microscopy for the presence of intact HFF. The average plaque areas (particles) were analyzed using ImageJ software.
Replication assay in T. gondii mutants
TgFLP12-HA-iKD mutants were pre-treated for 3 days with 0.5 μg/ml ATc before replication assay. In 4-well plates (with coverslips) equal numbers of parasites were allowed to invade confluent HFF cells for 2 h. After invasion, the plates were washed (x3) with ED1 (DMEM, 1% FBS) and parasites were grown for 24 h with or without ATc (0.5 μg/ml). These coverslips were then processed for IFA and the number of parasites per vacuole was calculated (n = 4 independent experiments).
Electron microscopy
TgFLP12-iKD parasites grew 48 h in the presence and absence of ATc, in labteks (Nunk, Thermofisher). The labteks containing parasite infected HFF were fixed in 0.1 M cacodylate buffer with 2.5% glutaraldehyde for 2 h and kept at 4 °C until further processing. During processing, samples were fixed again for 1 h with 1% osmium tetroxide in cacodylate buffer followed by overnight treatment in 2% uranyl acetate in distilled water. After dehydration in graded series of acetonitrile, samples were progressively impregnated in Epon812, the wells were then filled with fresh resin and allowed to polymerize for 48 h at 60 °C. Ultrathin 70 nm sections were obtained with a Leica UCT Ultramicrotome and collected on copper grids. Grids were poststained with uranyl acetate and lead citrate before their observation on a Jeol1200EXII Transmission Electron Microscope. All chemicals were from electron microscopy sciences.
Lipidomic analysis
To extract lipids, T. gondii tachyzoites were grown on 175 cm² HFF cultures. Freshly egressed parasites ( ~ 108 parasites equivalent) were quenched in a bath of pure ethanol/dry ice, parasites are pushed out their vacuoles after passage in 23 G needle, filtered, and counted by hemocytometer before centrifugation. Pellets were washed 3x times in cold PBS to eliminate lipids from the culture media before conservation at −80 °C. Internal standard 10 µmol FFA C13:0 and 10 µmol PC C21:0 (Avanti Polar lipids) are added to samples and lipids are extracted following the Bligh and Dyer modified by Folch protocol54, FAs are derivatized using Trimethylsulfonium hydroxide and analyzed by gas-chromatography mass-spectrometry (Agilent 5977A-7890B) (GC-MS), fatty acid methyl esters were identified by their mass spectrum and retention time compared to authentic standards.
Phospholipid, DAG/TAG/FFA analyses: The extracted total lipid extracted (as above) was separated by one-dimensional silica gel high-performance thin-layer chromatography (HPTLC, Merck). The 1st and 2nd solvent systems used were chloroform/methanol/water/ acetic acid, 25:15:2:4 (v/v/v/v) and hexane/MTBE/acetic acid, 35:15:0.5 (v/v/v), respectively. For DAG, TAG, FFA, and PL analysis, the total lipid fraction was separated by 1D-HPTLC using hexane/diethyl ether/formic acid, 80:20:2 (v/v/v) as solvent system. The spots correlating to DAG, TAG, FFA, and PL on the HPTLC plate were scraped off, and lipids were methanolized with 200 μL 0.5 M methanolic HCl in the presence of 1 nmol pentadecanoic acid (C15:0) as internal standard at 85 °C for 4 h. The resulting FAMEs were extracted with hexane and analyzed by GC-MS (Agilent).
Stable isotope labeling analysis
Confluent T175 HFF cultures were set up for FASII 13Carbon labeling, the culture medium was discarded and replaced with ED1% (-) Glucose complemented with a final concentration of 8 mM U-13C-Glucose or 8 mM U-12C-Glucose as a control for 13C natural abundance. Freshly egressed parasites are loaded on the previous HFF U-13C-Glucose / U-12C-Glucose and incubated for 72 h before parasites harvest and lipid extraction for GC-MS analysis. Scavenging was assessed as described in Dass et al. 6, T175 are set up with HFF grown for 5 days in D10% (-)Glucose complemented with a final concentration of 8 mM U-13C-Glucose. Confluent T175 HFF pre-labeled cultures are set up for scavenging assay, the culture medium is discarded, cells are washed with ED1% (3x) and new ED1% is added. Freshly egressed parasites are loaded on the previous HFF pre-labeled with U-13C-Glucose and incubated for 72 h before parasites harvest and lipid extraction for GC-MS analysis.
Heterologous complementation in yeast Δfat1 and deuterated FA import
The pRS426-MET25 vector served as the backbone for constructing a plasmid carrying the complete TgFLP12 cDNA sequence. The ORF was synthesized by GenScript, based on the predicted transcribed sequence of TgFLP12 obtained from ToxoDB.org. This sequence was codon-optimized for heterologous expression in yeast and cloned into the vector using EcoRI and KpnI restriction sites. All yeast strains were transformed either with this plasmid or with the empty vector (Ф) as a control. The yeast strains used in this study are listed in Table 2.
Cultures were grown in SC-URA medium with 0.2% glucose under standard conditions at 30 °C. For metabolic labeling, SC-URA was supplemented with either D31-C16:0 or D33-C18:1 deuterated FAs (Sigma) at 50 µM for 7 h. After being grown in the presence or absence of deuterated FA, the yeast cultures were pelleted, then metabolically quenched as previously described6, and processed for MS-based lipidomic analyses, see below. All fatty acid species, including deuterated FAs, were then quantified and subjected to statistical analyses to determine the FA import capacity of each yeast strain.
Growth assays were conducted in 96-well microplates (Thermo Scientific Nunc MicroWell, Nunclon Delta-Treated, Flat-Bottom Microplate) containing 200 µL of SC-URA per well. Plates were incubated at 30 °C, and optical density at 600 nm (OD600) was measured every 15 min for ~24 h.
Statistics and graphs
Graphs and statistical analysis were made using GraphPad Prism10 (10.4.1). Graphs represent means and error bars represent standard errors of means. All data were analyzed with the two-tailed unpaired student’s t test, except when mentioned in the legends. All p values lower than 0.05 are considered significant and indicated in the figures. All graphs are made from at least three biological replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the paper, its Supplementary Information, the Source data file provided with this manuscript, on the MetaboLights website (#REQ20250604210987), and/or can be obtained upon request to the corresponding author. Source data are provided with this paper.
References
Paris, L. 106 - Toxoplasmosis. in Hunter’s Tropical Medicine and Emerging Infectious Diseases (Tenth Edition) (eds. Ryan, E. T., Hill, D. R., Solomon, T., Aronson, N. E. & Endy, T. P.) 803–813 (Elsevier, 2020).
Mazumdar, J., Wilson, H. E., Masek, K., Hunter, A. C. & Striepen, B. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc. Natl Acad. Sci. USA 103, 13192–13197 (2006).
Pernas, L., Bean, C., Boothroyd, J. C. & Scorrano, L. Mitochondria restrict growth of the intracellular parasite toxoplasma gondii by limiting its uptake of fatty acids. Cell Metab. 27, 886–897.e4 (2018).
Nolan, S. J., Romano, J. D. & Coppens, I. Host lipid droplets: an important source of lipids salvaged by the intracellular parasite Toxoplasma gondii. PLoS Pathog. 13, e1006362 (2017).
Nolan, S. J., Romano, J. D., Kline, J. T. & Coppens, I. Novel approaches to kill Toxoplasma gondii by exploiting the uncontrolled uptake of unsaturated fatty acids and vulnerability to lipid storage inhibition of the parasite. Antimicrob. Agents Chemother. 62, e00347–18 (2018).
Dass, S. et al. Toxoplasma LIPIN is essential in channeling host lipid fluxes through membrane biogenesis and lipid storage. Nat. Commun. 12, 2813 (2021).
Shunmugam, S., Arnold, C.-S., Dass, S., Katris, N. J. & Botté, C. Y. The flexibility of Apicomplexa parasites in lipid metabolism. PLoS Pathog. 18, e1010313 (2022).
Wilson, R. J. et al. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J. Mol. Biol. 261, 155–172 (1996).
McFadden, G. I., Reith, M. E., Munholland, J. & Lang-Unnasch, N. Plastid in human parasites. Nature 381, 482–482 (1996).
Köhler, S. et al. A plastid of probable green algal origin in Apicomplexan parasites. Science 275, 1485–1489 (1997).
Waller, R. F. et al. Nuclear-encoded proteins target to the plastid in TOXOPLASMA GONDII AND PLASMOdium falciparum. Proc. Natl Acad. Sci. USA 95, 12352–12357 (1998).
Yu, M. et al. The fatty acid biosynthesis enzyme fabi plays a key role in the development of liver stage malarial parasites. Cell Host Microbe 4, 567–578 (2008).
Vaughan, A. M. et al. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell. Microbiol. 11, 506–520 (2009).
Amiar, S. et al. Apicoplast-localized lysophosphatidic acid precursor assembly is required for bulk phospholipid synthesis in toxoplasma gondii and relies on an algal/plant-like glycerol 3-phosphate acyltransferase. PLoS Pathog. 12, e1005765 (2016).
van Schaijk, B. C. L. et al. Type II fatty acid biosynthesis is essential for Plasmodium falciparum sporozoite development in the midgut of Anopheles mosquitoes. Eukaryot. Cell 13, 550–559 (2014).
Krishnan, A. et al. Functional and computational genomics reveal unprecedented flexibility in stage-specific toxoplasma metabolism. Cell Host Microbe 27, 290–306.e11 (2020).
Liang, X. et al. Acquisition of exogenous fatty acids renders apicoplast-based biosynthesis dispensable in tachyzoites of Toxoplasma. J. Biol. Chem. 295, 7743–7752 (2020).
Nakamura, Y., Tsuchiya, M. & Ohta, H. Plastidic phosphatidic acid phosphatases identified in a distinct subfamily of lipid phosphate phosphatases with prokaryotic origin*. J. Biol. Chem. 282, 29013–29021 (2007).
Amiar, S. et al. Division and adaptation to host environment of apicomplexan parasites depend on apicoplast lipid metabolic plasticity and host organelle remodeling. Cell Rep. 30, 3778–3792.e9 (2020).
Upadhyay, S. K. et al. Structural insights into the ACYL intermediates of the Plasmodium falciparum fatty acid synthesis pathway: the mechanism of expansion of the acyl carrier protein core. J. Biol. Chem. 284, 22390–22400 (2009).
Xu, X.-P. et al. The role of type II fatty acid synthesis enzymEs FabZ, ODSCI, and ODSCII in the pathogenesis of toxoplasma gondii infection. Front Microbiol 12, 703059 (2021).
Alonso, A. M. et al. Exploring protein myristoylation in Toxoplasma gondii. Exp. Parasitol. 203, 8–18 (2019).
Broncel, M. et al. Profiling of myristoylation in Toxoplasma gondii reveals an N-myristoylated protein important for host cell penetration. eLife 9, e57861 (2020).
Mullin, K. A. et al. Membrane transporters in the relict plastid of malaria parasites. Proc. Natl. Acad. Sci. USA 103, 9572–9577 (2006).
Sayers, C. P., Mollard, V., Buchanan, H. D., McFadden, G. I. & Goodman, C. D. A genetic screen in rodent malaria parasites identifies five new apicoplast putative membrane transporters, one of which is essential in human malaria parasites. Cell. Microbiol. 20, 12789 (2018).
Boucher, M. J. et al. Integrative proteomics and bioinformatic prediction enable a high-confidence apicoplast proteome in malaria parasites. PLoS Biol. 16, e2005895 (2018).
Kloehn, J., Lacour, C. E. & Soldati-Favre, D. The metabolic pathways and transporters of the plastid organelle in Apicomplexa. Curr. Opin. Microbiol. 63, 250–258 (2021).
Lu, B., Xu, C., Awai, K., Jones, A. D. & Benning, C. A small ATPase protein of arabidopsis, TGD3, involved in chloroplast lipid import. J. Biol. Chem. 282, 35945–35953 (2007).
Fan, J., Zhai, Z., Yan, C. & Xu, C. Arabidopsis trigalactosyldiacylglycerol5 Interacts with TGD1, TGD2, and TGD4 to facilitate lipid transfer from the endoplasmic reticulum to plastids. Plant Cell 27, 2941–2955 (2015).
Roston, R., Gao, J., Xu, C. & Benning, C. Arabidopsis chloroplast lipid transport protein TGD2 disrupts membranes and is part of a large complex: TGD2 involved in chloroplast lipid transport. Plant J. 66, 759–769 (2011).
Li, N. et al. FAX1, a novel membrane protein mediating plastid fatty acid export. PLoS Biol. 13, e1002053 (2015).
Li, N. et al. Two plastid fatty acid exporters contribute to seed oil accumulation in arabidopsis1. Plant Physiol. 182, 1910–1919 (2020).
Palmgren, M. G. & Nissen, P. P-type ATPases. Annu Rev. Biophys. 40, 243–266 (2011).
Nemirovskaya, T. L. & Sharlo, K. A. Roles of ATP and SERCA in the regulation of calcium turnover in unloaded skeletal muscles: current view and future directions. Int J. Mol. Sci. 23, 6937 (2022).
Bullen, H. E. et al. Phosphatidic acid-mediated signaling regulates microneme secretion in toxoplasma. Cell Host Microbe 19, 349–360 (2016).
Günay-Esiyok, Ö & Gupta, N. Chimeras of P4-ATPase and guanylate cyclase in pathogenic protists. Trends Parasitol. 36, 382–392 (2020).
Günay-Esiyok, Ö, Scheib, U., Noll, M. & Gupta, N. An unusual and vital protein with guanylate cyclase and P4-ATPase domains in a pathogenic protist. Life Sci. Alliance 2, e201900402 (2019).
Katris, N. J. et al. Rapid Kinetics of Lipid Second Messengers Controlled by a cGMP Signalling Network Coordinates Apical Complex Functions in Toxoplasma Tachyzoites. http://biorxiv.org/lookup/doi/10.1101/2020.06.19.160341 (2020).
Bisio, H., et al. Toxoplasma gondii phosphatidylserine flippase complex ATP2B-CDC50.4 critically participates in microneme exocytosis. PLoS Pathog. 18, e1010438 (2022)
Bisio, H., Lunghi, M., Brochet, M. & Soldati-Favre, D. Phosphatidic acid governs natural egress in Toxoplasma gondii via a guanylate cyclase receptor platform. Nat. Microbiol. 4, 420–428 (2019).
Nagamune, K., Moreno, S. N., Chini, E. N. & Sibley, L. D. Calcium regulation and signaling in apicomplexan parasites. Subcell. Biochem. 47, 70–81 (2008).
Holpert, M., Gross, U. & Bohne, W. Disruption of the bradyzoite-specific P-type (H+)-ATPase PMA1 in Toxoplasma gondii leads to decreased bradyzoite differentiation after stress stimuli but does not interfere with mature tissue cyst formation. Mol. Biochem. Parasitol. 146, 129–133 (2006).
Chen, K. et al. Aminoglycerophospholipid flipping and P4-ATPases in Toxoplasma gondii. J. Biol. Chem. 296, 100315 (2021).
McKenna, M. J. et al. The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Science 369, 5809 (2020).
Li, T. et al. P5A ATPase controls ER translocation of WNT in neuronal migration. Cell Rep. 37, 109901 (2021).
De La Hera, D. P., Corradi, G. R., Adamo, H. P. & De Tezanos Pinto, F. Parkinson’s disease-associated human P5B-ATPase ATP13A2 increases spermidine uptake. Biochem. J. 450, 47–53 (2013).
Van Veen, S., Martin, S., Schuermans, M. & Vangheluwe, P. Polyamine transport assay using reconstituted yeast membranes. Bio Protoc. 11, e3888 (2021).
Barylyuk, K. et al. A comprehensive subcellular atlas of the toxoplasma proteome via hyperlopit provides spatial context for protein functions. Cell Host Microbe 28, 752–766 (2020).
Huynh, M.-H. & Carruthers, V. B. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell 8, 530–539 (2009).
Chen, P. et al. A pyruvate transporter in the apicoplast of apicomplexan parasites. Proc. Natl. Acad. Sci. USA 121, e2314314121 (2024).
Sidik, S. M. et al. A Genome-wide CRISPR screen in toxoplasma identifies essential apicomplexan genes. Cell 166, 1423–1435.e12 (2016).
ToxoDB. https://toxodb.org/toxo/app.
Walsh, D., Katris, N. J., Sheiner, L. & Botté, C. Y. Toxoplasma metabolic flexibility in different growth conditions. Trends Parasitol. 38, 775–790 (2022).
Ramakrishnan, S. et al. Apicoplast and endoplasmic reticulum cooperate in fatty acid biosynthesis in apicomplexan parasite toxoplasma gondii*. J. Biol. Chem. 287, 4957–4971 (2012).
Dong, H. et al. The Toxoplasma monocarboxylate transporters are involved in the metabolism within the apicoplast and are linked to parasite survival. Elife 12, RP88866 (2024).
Renaud, E. A. et al. Disrupting the plastidic iron-sulfur cluster biogenesis pathway in Toxoplasma gondii has pleiotropic effects irreversibly impacting parasite viability. J. Biol. Chem. 298, 102243 (2022).
Lévêque, M. F. et al. TgPL2, a patatin-like phospholipase ___domain-containing protein, is involved in the maintenance of apicoplast lipids homeostasis in Toxoplasma. Mol. Microbiol. 105, 158–174 (2017).
Dubois, D. et al. Toxoplasma gondii acetyl-CoA synthetase is involved in fatty acid elongation (of long fatty acid chains) during tachyzoite life stages. J. Lipid Res. 59, 994–1004 (2018).
Charital, S. et al. The acyl-CoA synthetase TgACS1 allows neutral lipid metabolism and extracellular motility in Toxoplasma gondii through relocation via its peroxisomal targeting sequence (PTS) under low nutrient conditions. mBio 15, e0042724 (2024).
Dass, S. et al. Toxoplasma acyl-CoA synthetase TgACS3 is crucial to channel host fatty acids in lipid droplets and for parasite propagation. J. Lipid Res. 65, 100645 (2024).
Ramakrishnan, S. et al. The intracellular parasite Toxoplasma gondii depends on the synthesis of long chain and very long-chain unsaturated fatty acids not supplied by the host cell. Mol. Microbiol. 97, 64–76 (2015).
Cook, T. et al. Divergent polyamine metabolism in the Apicomplexa. Microbiology 153, 1123–1130 (2007).
Faergeman, N. J., DiRusso, C. C., Elberger, A., Knudsen, J. & Black, P. N. Disruption of the Saccharomyces cerevisiae homologue to the murine fatty acid transport protein impairs uptake and growth on long-chain fatty acids. J. Biol. Chem. 272, 8531–8538 (1997).
DiRusso, C. C. et al. Comparative biochemical studies of the murine fatty acid transport proteins (FATP) expressed in yeast. J. Biol. Chem. 280, 16829–16837 (2005).
Zou, Z., DiRusso, C. C., Ctrnacta, V. & Black, P. N. Fatty acid transport in Saccharomyces cerevisiae. Directed mutagenesis of FAT1 distinguishes the biochemical activities associated with Fat1p. J. Biol. Chem. 277, 31062–31071 (2002).
Lu, B. & Benning, C. A 25-amino acid sequence of the arabidopsis TGD2 protein is sufficient for specific binding of phosphatidic acid. J. Biol. Chem. 284, 17420–17427 (2009).
https://piroplasmadb.org/piro/app/ PiroplasmaDB.
Brayton, K. A. et al. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathog. 3, 1401–1413 (2007).
Yeh, E. & DeRisi, J. L. Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage plasmodium falciparum. PLoS Biol. 9, e1001138 (2011).
Quittnat, F. et al. On the biogenesis of lipid bodies in ancient eukaryotes: synthesis of triacylglycerols by a Toxoplasma DGAT1-related enzyme. Mol. Biochem. Parasitol. 138, 107–122 (2004).
Sanchez, S. G. et al. The apicoplast is important for the viability and persistence of Toxoplasma gondii bradyzoites. Proc. Natl. Acad. Sci. USA. 120, e2309043120 (2023).
Axelsen, K. B. & Palmgren, M. G. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46, 84–101 (1998).
Benito, B., Garciadeblás, B., Schreier, P. & Rodríguez-Navarro, A. Novel p-type ATPases mediate high-affinity potassium or sodium uptake in fungi. Eukaryot. Cell 3, 359–368 (2004).
Rodríguez-Navarro, A. & Benito, B. Sodium or potassium efflux ATPase a fungal, bryophyte, and protozoal ATPase. Biochim. Biophys. Acta 1798, 1841–1853 (2010).
Acknowledgements
This work was supported by Agence Nationale de la Recherche, France (Project ApicoLipiAdapt grant ANR-21-CE44-0010; Project Apicolipidtraffic grant ANR-23-CE15-0009-01; Project OIL grant ANR-24-CE15-2171-02), The Fondation pour la Recherche Médicale (FRM EQU202103012700), Laboratoire d’Excellence Parafrap, France (grant ANR-11-LABX-0024), LIA-IRP CNRS and INSERM Programs (Apicolipid projects), the Université Grenoble Alpes (IDEX ISP Apicolipid) and Région Auvergne Rhone-Alpes for the lipidomics analyses platform (Grant IRICE Project GEMELI), Collaborative Research Program Grant CEFIPRA (Project 6003-1) by the CEFIPRA (MESRI-DBT) to CYB. JJ was supported by the Junior STAR grant no. 21-19798 M from the Czech Science Foundation.
Author information
Authors and Affiliations
Contributions
C.Y.B., Y.Y.B., N.J.K. and C.S.A. contributed to the design of the work. C.Y.B., Y.Y.B., N.J.K. and C.S.A. contributed to the interpretation of data. C.S.A., S.S., S.D., S.C., J.J., L.B., D.J., C.L.V., P.C., A.M.A., J.G., T.G., M.F.C., N.J.K., Y.Y.B. and C.Y.B. contributed to the acquisition and analysis of data. C.Y.B. designed, organized, led and financed the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Arnold, CS., Alazzi, AM., Shunmugam, S. et al. A P5-ATPase, TgFLP12, diverging from plant chloroplast lipid transporters mediates apicoplast fatty export in Toxoplasma. Nat Commun 16, 5538 (2025). https://doi.org/10.1038/s41467-025-61155-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-61155-9