Abstract
Variants in GBA1 result in dysregulated sphingolipids. We investigated five CSF d18:1 sphingolipid species in a longitudinal multicenter cohort comprising people with Parkinson’s Disease and Dementia with Lewy bodies with and without GBA1 variants and healthy controls. We found no increase of sphingolipid species in heterozygous GBA1 variant participants and no effect on development of cognitive impairment. Thus, CSF d18:1 sphingolipids are not suitable as state markers in Parkinson’s Disease.
Similar content being viewed by others
Introduction
Bi-allelic variants in the glucocerebrosidase gene (GBA1) cause Gaucher Disease (GD). Heterozygous variants in GBA1 represent the most important genetic risk factor for alpha-synucleinopathies with Lewy-body pathology, Parkinson’s Disease (PD), and Dementia with Lewy Bodies (DLB)1. GBA1 encodes the lysosomal enzyme glucocerebrosidase (GCase), which metabolizes glucosylceramides (GlcCer) and glucosylsphingosines (GlcSph). Experimental evidence suggests that GBA1 variants result in lower GCase activity and cause a build-up of GlcCer2 which also impairs lysosomal function and the degradation of alpha-synuclein (α-synuclein)3. In addition to these directly linked metabolites, downstream products of the broader sphingolipid pathway (including sphingosine (Sph) and sphingosine-1-phosphate (S1P)) seem also affected due to GBA1 variants4. Data from α-synuclein/GBA1 transgenic mice and cell cultures support this notion indicating that not only GlcCer but also GlcSph, Sph, and S1P promote the aggregation of α-synuclein5,6,7. Notably, GlcSph has been shown to cause α-synuclein oligomerization and aggregation even more prominently than GlcCer6. The link between GBA1 with PD and DLB offers various therapeutic targets which, in turn, require reliable (fluid)biomarkers to monitor trait, state, target engagement, and/or response to therapy. In GD, GlcCer, although it is the direct substrate and more abundant than GlcSph, is not a useful biomarker8. In contrast, plasma levels of GlcSph show good correlation with disease severity and are used to monitor response to therapy. A recent study reported elevated plasma levels of GlcSph d18:1 isoform in participants with and without PD carrying a heterozygous GBA1 N370S variant compared to PD and healthy participants without pathogenic GBA1 or LRRK2 G2019S variants9. All other assessed sphingolipids (total GlcCer, Cer, Galactosylceramide (GalCer), and Galactosylsphingosine (GalSph)) showed similar mean plasma levels between groups. While this finding is interesting, several key questions remain unanswered: 1. Is this peripheral GlcSph signature representative of the central nervous system and similarly found in CSF of heterozygous GBA1 variant carriers? 2. The N370S variant is one of several important GBA1 variants. How do sphingolipid profiles look in other GBA1 variant groups? 3. Apart from PD, heterozygous GBA1 variants are prominently associated with DLB. Are sphingolipid profiles found in PD similar or even more prominent in DLB participants carrying GBA1 variants as both entities represent a biological continuum? To address these questions, we investigated 5 CSF d18:1 sphingolipid species (Cer (d18:1/18:0), GlcCer (d18:1/18:0), SphM (d18:1/18:0), GlcSph (d18:1) and GalSph (d18:1)) in a multicenter cohort with 152 PD and 37 DLB participants with and without heterozygous GBA1 variants. We also included 5 samples from PD participants who were bi-allelic carriers of GBA1 variants as “positive controls” and 40 healthy elderly as control participants. We estimated the effect of sphingolipid CSF levels on longitudinal development of cognitive impairment in PD.
CSF levels of d18:1 sphingolipids by GBA1 genotype and GBA1 severity
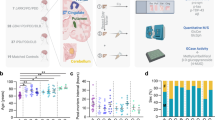
We combined PD and DLB groups and stratified by GBA1 genotype. Both, PDGBA + DLBGBA as well as PDGBA_WT + DLBGBA_WT groups had lower levels of GalSph compared to HC. All other assessed sphingolipid species did not differ between GBA1 and WT status; Table 1.
Subanalysis of PD and DLB stratified by GBA1 variant severity (GBAWT, GBArisk, GBAmild, GBAsevere) revealed no significant differences in levels of sphingolipid species in pmol/ml (Cer: 3.76 vs. 4.18 vs. 3.86 vs. 3.65; GlcCer: 0.90 vs. 0.94 vs. 0.92 vs. 0.86; GlcSph: 0.013 vs. 0.014 vs. 0.015 vs. 0.014; GalSph: 0.15 vs. 0.16 vs. 0.15 vs. 0.13; SphM: 333 vs. 346 vs. 346 vs. 324; p > 0.05 respectively); Supplemental Fig. 1.
CSF levels of d18:1 sphingolipids by GBA1 genotype and disease group
DLBGBA_WT had lower levels of Cer than PDGBA, PDGBA_WT, and HC (p < 0.05, respectively). DLBGBA and DLBGBA_WT had lower levels of GalSph than HC (p < 0.05 and p < 0.01). Overall, but purely numerically, DLBGBA had higher levels of all sphingolipid species compared to DLBGBA_WT.
In PD, levels of GlcSph were numerically higher in PDGBA compared to PDGBA_WT while levels of all other sphingolipid species showed no clear trend.
As proof-of-concept, PDGaucher by far showed the highest levels of GlcSph (4-fold increase) compared to all other groups; Fig. 1.
DLBGBA_WT patients had lower levels of ceramide than HC, PDGBA_WT, and PDGBA patients (A). PDGaucher patients had higher levels of glucosylsphingosine than HC, PDGBA_WT, PDGBA, DLBGBA_WT, and DLBGBA (C). DLBGBA_WT and DLBGBA patients had lower levels of galactosylsphingosine than HC (D). There is no difference in levels of glucosylceramide and sphingomyelin between the patient groups (B + E). The red line indicates median levels of healthy controls, respectively. *p < 0.05; **p < 0.01;***p < 0.001.
Correlation between CSF d18:1 sphingolipids and clinical measurements
In PDGBA_WT + DLBGBA_WT, higher levels of GalSph (ρ = −0.276, p = 0.005) were associated with lower H&Y. There were no significant correlations between sphingolipid species and clinical scores (H&Y, UPDRS-III and MoCA) in the combined group of GBA1 variant carriers PDGBA + DLBGBA. There were no correlations between levels of sphingolipid species and GBA1 severity.
Longitudinal association of baseline CSF d18:1 sphingolipid tertiles with cognitive impairment
Kaplan-Meier analyses revealed no differences in reaching the milestone cognitive impairment when comparing the lowest with the highest tertile of any sphingolipid species, neither in the combined group of all PD patients nor when analysing tertiles of PDGBA and PDGBA_WT separately; Supplemental Fig. 2.
By measuring targeted CSF d18:1 isoforms of various sphingolipid species (d18:1 GlcSph, GalSph, d18:1, 18:0 Cer, GlcCer, and SphM) in a multi-center cohort comprising 152 PD and 37 DLB participants with and without various heterozygous GBA1 variants and 40 HC we found no clear increase of the key substrates GlcCer and GlcSph when comparing GBA1 variant carriers with their WT comparator group. Notably, levels in PD and DLB with and without GBA1 variant were within the range of healthy elderly. Although PDGBA and DLBGBA had numerically higher mean levels of GlcSph compared to PDGBA_WT and DLBGBA_WT, these small differences seem not adequate for stratification of GBA1 variant carriers to enter clinical trials. Thereby, our present CSF data do not robustly support findings from the recent study in plasma which reported significantly elevated levels of d18:1 GlcSph in participants with and without PD carrying a heterozygous GBA1 N370S variant compared to PD and healthy participants without GBA1 variant9. It is noteworthy that we previously showed higher CSF levels of total GlcCer in PDGBA compared to PDGBA_WT in two different cohorts (Tuebingen GBA1 cohort with 50% overlap with the present Tuebingen cohort, PPMI cohort)4. The different analytical approaches might partially explain these discrepancies. In the present study, we focused on the d18:1, 18:0 isoform using a targeted analysis with internal spiked heavy-labeled standards that enable absolute quantification with the main goal to be used as endpoints for treatment trials. Opposed to that, we used untargeted lipidomics and summed up all measured isoforms of each sphingolipid species (total Cer, total GlcCer) for the previous analysis. At that time, we identified higher CSF levels of Sph in PDGBA compared to PDGBA_WT, but we did not measure GlcSph. Moreover, increased levels of total GlcCer in plasma have been reported in PD versus HC using a HPLC-based method, again summarizing all measured isoforms10. It was recently speculated that the very long-chain isoforms such as 22:0, 22:1, 23:0, 23:1, 24:0, and 24:1 are higher in GBA1 and thereby contribute to the significance when comparing sum levels of all isoforms. While using untargeted lipidomics provides good coverage of various sphingolipid chain lengths it has lower sensitivity and only provides relative quantification. We, therefore, suggest to explore the very long-chain 22-24 isoforms in a targeted approach with absolute quantification in future studies.
We did not identify relevant correlations between sphingolipid levels and relevant clinical scores (UPDRS-III, HY, MoCA) nor did we detect differences between any sphingolipid tertile group with longitudinal development of cognitive impairment, neither in the combined group of all PD patients nor when analysing tertile groups of PDGBA and PDGBA_WT separately. These findings indicate that CSF levels of sphingolipids are not suited to monitor disease burden or progression in PD. This is further supported by the finding that DLBGBA do not show higher sphingolipid levels than PDGBA. These findings are in line with the recent study in plasma where no association was found between plasma levels of any of the assessed sphingolipid species and PD disease status9. Thereby, and opposed to GD, (d18:1, 18:0) sphingolipid isoforms and especially GlcSph (d18:1) are not useful as state markers in PD.
The assay’s validity seems reasonable with a 4-fold increase of CSF GlcSph levels in PDGD. While the additional inclusion of DLB participants and the inclusion of different GBA1 variants pose strengths of the present analyses, we acknowledge the following limitations: (I) The measurement of targeted single sphingolipid isoform (d18:1, 18:0) instead of several isoforms limits the detection of effects coming from long-chain isoforms. (II) Longitudinal measurements of sphingolipids in de-novo and enriched at-risk individuals are important to evaluate changes over time, with aging and/or with disease progression. (III) Blood-CSF pairs from the same individual at the same time point are needed. (IV) It is worth mentioning that even in the bi-allelic participants CSF levels of Cer, GlcCer, SphM, and GalSph were low in general. It would be interesting to compare these levels to those from Gaucher disease patients without PD in future studies.
Methods
Participants
Together, the four sites (Karolinska University Hospital, Stockholm, Sweden; IRCCS Mondino Foundation of Pavia, Italy; University of Tuebingen, Germany; Vall d’Hebron Research Institute, Barcelona, Spain) collected CSF of 79 idiopathic PD patients (PDGBA_WT), 73 PD patients with GBA1 variant (PDGBA), 26 idiopathic DLB patients (DLBGBA_WT), 11 DLB patients with GBA1 variants (DLBGBA) and 5 PDGaucher patients. Forty neurodegenerative healthy elderly (spouses, volunteers) served as control individuals (HC).
Clinical investigations
Diagnosis of PD was defined according to UK Brain Bank Society Criteria or MDS clinical diagnostic criteria and diagnosis of DLB according to the DLB consortium revised consensus criteria. All participants were categorized by the modified Hoehn and Yahr Scale (H&Y), assessed with the Unified Parkinson’s Disease Rating Scale (UPDRS-III), and Montreal Cognitive Assessment (MoCA) and/or the Mini-Mental Status Examination (MMSE). Since the MoCA was available only from 2009 on, all previously obtained MMSE scores were converted into MoCA equivalent scores according to a published algorithm11. Presence of cognitive impairment was defined as MoCA score < 26. For demographic and clinical details see Supplemental Table 1.
Genetic screening
Genetic screening of the Tuebingen cohort for GBA1 variants was done by Sanger sequencing of all exons in 58% of the PD and 100% of DLB patients. In 42% of PD patients primary genetic screening was done by NeuroChip and in case of GBA1 variants confirmation by Sanger sequencing.
Genetic screening for the Stockholm cohort was performed with pyrosequencing and TaqMan PCR with subsequent confirmation by Sanger sequencing.
Genetic screening of the majority of the Italian cohort was performed by an NGS-based method. In a minority of patients, the GBA1 gene was tested by Sanger sequencing. Identified variants were all validated by Sanger sequencing.
Genetic screening for the Vall d’Hebron Initiative for Parkinson (VHIP) cohort was performed by an NGS-based method that combines a primary GBA1-specific long-range PCR with subsequent GBA1-exon-specific PCR and next-generation sequencing of the resulting products.
GBA1-subgroup classification of variant severity was based on established genotype risks reported for PD (PDGBA_severe, PDGBA_mild, PDGBA_risk).
An overview of GBA1 variants is given in Supplemental Table 2.
CSF measurement of sphingolipids
CSF Hb was measured using a sensitive ELISA assay (Bethyl Hb assay kit, Fortis Life Sciences, USA). Samples with Hb levels > 50 ng/mL were excluded in analysis.
Five CSF sphingolipid species (Cer (d18:1/18:0), GlcCer (d18:1/18:0), SphM (d18:1/18:0), GlcSph (d18:1) and GalSph (d18:1)) were measured using LC-MS/MS method at Ardena Bioanalysis BV, NL. The d18:1/18:0 variants of the specific sphingolipids were assessed since these are highly expressed in brain12,13. The analytical ranges were 0.500–50.0, 0.250–25.0, 25.0–2500, 0.00500–1.00, and 0.005–1.00 pmol/mL, respectively. All reference materials were obtained from Avanti Polar Lipids Inc., Alabaster AL, USA). All samples were measured blinded to clinical and genetic status.
Cer (d18:1/18:0) and spiked internal standard Cer (d18:1-d7/18:0) were isolated from 50 µL human CSF by protein precipitation with acetonitrile (ACN): isopropanol (IPA) (60:40 v/v). After precipitation, blank-, calibration-, QC-, and study samples were injected into the Chromatographic system (Shimadzu Nexera, Kyoto, Japan) on an xSelect CSH Phenyl-Hexyl column (100 × 3.0 mm (length × internal diameter), 2.5 µm (particle size), Waters, Milford, MA, USA) using isocratic elution with water: IPA:ACN:FA (formic acid) (16.5:18.5:65:0.1 (v/v/v/v)) as mobile phase. An API6500 tandem mass spectrometer (Sciex, Framingham, MA, USA) equipped with a Turbo Ion Spray probe operating in the positive multiple reaction monitoring mode was used for quantification. MS transitions were 566 → 264 and 573 → 271 (unit resolution) for Cer (d18:1/18:0) and Cer (d18:1-d7/18:0), respectively. The analytical range was 0.500–50.0 pmol/mL. Each analytical run included duplicate QC samples at three levels (QC-Low at 1.50 pmol/mL, QC-Medium at 8.00 pmol/mL, and QC-High at 40.0 pmol/mL) and a representative QC-Pool sample in quadruplicate.
GlcCer (d18:1/18:0) and SM (d18:1/18:0) and spiked internal standards GlcCer (d18:1-d5/18:1) and SM (d18:1/17:0) were isolated from 50 µL human CSF by liquid-liquid extraction (LLE) with ethyl acetate: IPA: Pentane (51:9:40 (v/v/v)). After extraction, blank-, calibration-, QC-, and study samples were injected into the Chromatographic system (Shimadzu) on a Hypersil GOLD Silica column (100 × 2.1 mm (length × internal diameter), 1.9 µm (particle size), Thermo Fisher Scientific, Waltham, MA, USA) using a gradient elution with 1 M NH4FA (ammonium formate) : FA:water: ACN (20:6:40:2000 (v/v/v/v)) as mobile phase A and 1 M NH4FA:water:ACN (5:500:10 (v/v/v)) as mobile phase B. An API6500 tandem mass spectrometer (Sciex) equipped with a Turbo Ion Spray probe operating in the positive multiple reaction monitoring mode was used for quantification. MS transitions for the analytes were 728 → 264 and 731 → 184 (unit resolution) for GlcCer (d18:1/18:0) and SM (d18:1/18:0), respectively. Transitions for the internals standards were 731 → 269 and 717 → 184, respectively. The analytical ranges were 0.250–25.0 and 25.0–2500 pmol/mL, respectively. Each analytical run included duplicate QC samples at three levels (QC-Low at 0.750/75.0 pmol/mL, QC-Medium at 4.00/400 pmol/mL, and QC-High at 20.0/2000 pmol/mL for GlcCer (d18:1/18:0) and SM (d18:1/18:0), respectively) and a representative QC-Pool sample in quadruplicate.
GlcSph (d18:1) and GalSph (d18:1) and spiked internal standards GlcSph (d18:1)-d5 and GalSph (d18:1)-d5 were isolated from 200 µL of human CSF by solid phase extraction (SPE) using 1cc 30 mg Waters Oasis MCX cartridges (Waters). In short, cartridges were equilibrated with 1 mL methanol (MeOH) and 1 mL water, after which the acidified CSF samples (0.2 mL CSF + 0.1 mL 1.0/2.0 mg/mL bovine serum albumin/ascorbic acid in phosphate buffer saline (PBS) + 0.75 mL 1% H3PO4 + 0.1 mL 2.0 mg/mL ascorbic acid in MeOH) were loaded on the cartridges. Sample were washed with 2 ×0.75 mL water and 2 × 0.75 mL MeOH after which the analytes were eluted with 2 × 0.4 mL 1% NH4FA in MeOH in a tube containing 0.1 mL of 2 mg/mL ascorbic acid in MeOH. After evaporation, extracts were redissolved in 0.0500 mL of 2 mg/mL ascorbic acid in chloroform:MeOH:UPW (5:1:0.1 (v/v/v)) after which 0.100 mL ACN was added. Blank-, calibration-, QC- and study samples were injected into the Chromatographic system (Shimadzu) on a Hypersil GOLD Silica column (100 × 2.1 mm (length × internal diameter), 1.9 µm (particle size)), Thermo Fisher Scientific) using a gradient elution with 1 M NH4FA:FA:water:ACN (20:6:40:2000 (v/v/v/v) as mobile phase A and 1 M NH4FA:water:ACN (5:500:10 (v/v/v) as mobile phase B. An API6500 tandem mass spectrometer (Sciex) equipped with a Turbo Ion Spray probe operating in the positive multiple reaction monitoring mode was used for quantification. MS transitions were 462 → 282 and 467 → 287 (unit resolution) for GlcSph (d18:1)/GalSph(d18:1) and GlcSph(d18:1)-d5/GalSph(d18:1)-d5, respectively. GlcSph(d18:1) and GalSph(d18:1) were chromatographically separated since the molecules are isomers and share similar fragments. The analytical ranges were 0.00500–1.00 pmol/mL for both analytes. Each analytical run included duplicate QC samples at three levels (QC-Low at 0.0300 pmol/mL, QC-Medium at 0.150 pmol/mL, and QC-High at 0.800 pmol/mL for both analytes) and a representative QC-Pool sample in quadruplicate.
Calibration samples were prepared fresh on the day of analysis in analyte free surrogate matrix using nine non-zero concentrations and at least one double blank (no analyte and no IS) and one blank (no IS). The calibration curves were calculated using a quadratic regression model and 1/x2 weighting. Results were accepted if the run met the predefined acceptance criteria as per FDA and EMA guidelines (1. Guidance for Industry: Bioanalytical Method Validation, U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER), Center for Veterinary Medicine (CVM), May 2018, BP. 2. Guideline on bioanalytical method validation, European Medicines Agency (EMA), Committee for Medicinal Products for Human Use (CHMP), EMEA/CHMP/EWP/192217/2009, 21 July 2011). For chromatograms of GluSph and GalSph please see supplemental material.
Statistical analysis
Statistical analysis was performed using SPSS 26.0 (IBM). Group comparisons of continuous data were analyzed using ANOVA/ANCOVA including age as co-variate where appropriate. The five PDGaucher were included as “positive controls” for a purely descriptive comparison (not powered for robust statistical analysis).
Pearson’s correlation was used to evaluate associations between CSF levels of sphingolipids and clinical scores. As PD and DLB are a biological continuum and to explicitly asses the GBA effect we combined the two wildtype groups (PDGBA_WT + DLBGBA_WT) and the two GBA1 variant groups (PDGBA + DLBGBA). As this study was exploratory, we did not correct for multiple testing. However, only correlations with at least a correlation coefficient of ρ > 0.20 were considered meaningful (irrespective if the p-value was <0.05).
For longitudinal analysis of association between CSF levels of sphingolipids and the GBA1-related key milestone cognitive impairment, we calculated Kaplan-Meier survival curves by stratifying all PD patients into sphingolipid tertiles based on the individual CSF level of the respective sphingolipid species. Kaplan-Meier survival curves were analyzed separately for PDGBA_WT and PDGBA.
Ethical approval and patient consent
The study was approved by the local Ethics Committees (Tuebingen:199/2011BO1; Pavia: PD-GEN, 16/01/2019 CE PV; Stockholm: 2016/19-31/2; Barcelona: PR(AG)170-2015, PR(AG)434-2019; ESCAPE Bio: Stitching Beoordeling Ethiek Biomedisch Onderzoek Review Board Assen, Netherlands). All participants gave written informed consent.
Data availability
Raw data of the measurements were uploaded to Zenodo: DOI 10.5281/zenodo.13132385. Further anonymized data are available upon request to: [email protected].
References
Sidransky, E. et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N. Engl. J. Med. 361, 1651–1661, https://doi.org/10.1056/NEJMoa0901281 (2009).
Beutler, E. Gaucher disease: new molecular approaches to diagnosis and treatment. Science 256, 794–799 (1992).
Mazzulli, J. R. et al. Gaucher disease glucocerebrosidase and alpha-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell 146, 37–52 (2011).
Lerche, S. et al. The mutation matters: CSF profiles of GCase, sphingolipids, alpha-synuclein in PD(GBA). Mov. Disord. 36, 1216–1228 (2021).
Taguchi, Y. V. et al. Glucosylsphingosine promotes alpha-synuclein pathology in mutant GBA-associated Parkinson’s disease. J. Neurosci. 37, 9617–9631 (2017).
Zunke, F. et al. Reversible conformational conversion of alpha-synuclein into toxic assemblies by glucosylceramide. Neuron 97, 92–107 e110 (2018).
Suzuki, M., Sango, K., Wada, K. & Nagai, Y. Pathological role of lipid interaction with alpha-synuclein in Parkinson’s disease. Neurochem. Int. 119, 97–106 (2018).
Barton, N. W. et al. Replacement therapy for inherited enzyme deficiency–macrophage-targeted glucocerebrosidase for Gaucher’s disease. N. Engl. J. Med. 324, 1464–1470 (1991).
Surface, M. et al. Plasma glucosylsphingosine in GBA1 mutation carriers with and without Parkinson’s disease. Mov. Disord. 37, 416–421 (2022).
Te Vruchte, D. et al. Glycosphingolipid changes in plasma in Parkinson’s disease independent of glucosylceramide levels. Mov. Disord. 37, 2129–2134 (2022).
Fasnacht, J. S. et al. Conversion between the Montreal cognitive assessment and the mini-mental status examination. J. Am. Geriatr. Soc. 71, 869–879 (2023).
Filippov, V. et al. Increased ceramide in brains with Alzheimer’s and other neurodegenerative diseases. J. Alzheimers Dis. 29, 537–547 (2012).
Olsen, A. S. B. & Faergeman, N. J. Sphingolipids: membrane microdomains in brain development, function and neurological diseases. Open Biol. 7 https://doi.org/10.1098/rsob.170069 (2017).
Harris, P. A. et al. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inf. 42, 377–381 (2009).
Acknowledgements
We would like to thank Mari Mauer, Dr. Enchi Liu, Dr. Marissa Gu, Dr. Maria Camprodón, and Jordi Riera for developing the CSF collection procedures and initiating the assay qualification work. The authors thank all participants and their families for their generosity. The Tübingen cohort was supported by BMBF (PDdementia) and DZNE (PD research; MIGAP study). Moreover, it was funded by the PD-Strat project (FKZ 031L0137B) which was supported by the German Federal Ministry of Education and Research (BMBF) in the frame of ERACoSysMed2. Study data were collected and managed using REDCap electronic data capture tools hosted at University of Tuebingen14. We acknowledge support by Open Access Publishing Fund of University of Tübingen. The Pavia cohort was supported by the Italian Ministry of Health—Ricerca Corrente 2018-2021 to IRCCS Mondino Foundation. The Stockholm cohort was supported by the Stockholm City Council and Knut and Alice Wallenbergs Foundation. The VHIP cohort was supported by the Fondo de Investigación Sanitaria-Instituto de Salud Carlos III (Spain)-FEDER (PI20/00728 and PI21/01603). Samples from patients in the VHIP cohort were provided by the Vall d’Hebron University Hospital Biobank (PT20/00107), integrated in the Spanish National Biobanks Network, and they were processed following standard operating procedures.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
S.L.: Coordination, data analysis, and interpretation, manuscript preparation. I.W.: Execution of the study (clinical and lumbar puncture), critical review of the manuscript. E.M.V.: Execution of the study (clinical and genetic testing), critical review of the manuscript. M.A.: Execution of the study (clinical and lumbar puncture), critical review of the manuscript. D.S.: Execution of the study (clinical and lumbar puncture), critical review of the manuscript. M.M.-V.: Execution of the study (genetic testing), critical review of the manuscript. J.H.-V.: Execution of the study (clinical and lumbar puncture), critical review of the manuscript. A.L.: Execution of the study (coordination), critical review of the manuscript. A.S.: Execution of the study (data collection), critical review of the manuscript. P.S.: Execution of the study (clinical and lumbar puncture), critical review of the manuscript. N.P.F.: Execution of the clinical study with CSF lumbar puncture and serial CSF sampling in healthy volunteers. C.B.: Execution of the clinical study with CSF lumbar puncture and serial sampling in healthy volunteers and oversight of CSF sphingolipid assay qualification. S.S.: Conceived the study collaboration, CSF sphingolipid assay qualification, bioanalysis of study samples, data analysis, interpretation of data, and manuscript preparation. K.B.: Conception, design, and execution of the study (clinical and lumbar puncture), interpretation of data, statistical analysis, manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
Authors S.S., N.P.F., and C.B. were employees of ESCAPE Bio at the time this work was performed. The remaining authors declare that the research was conducted in the absence of any commercial, financial, or non-financial relationships that relate to a potential conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lerche, S., Wurster, I., Valente, E.M. et al. CSF d18:1 sphingolipid species in Parkinson disease and dementia with Lewy bodies with and without GBA1 variants. npj Parkinsons Dis. 10, 198 (2024). https://doi.org/10.1038/s41531-024-00820-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41531-024-00820-0